Figure 2.
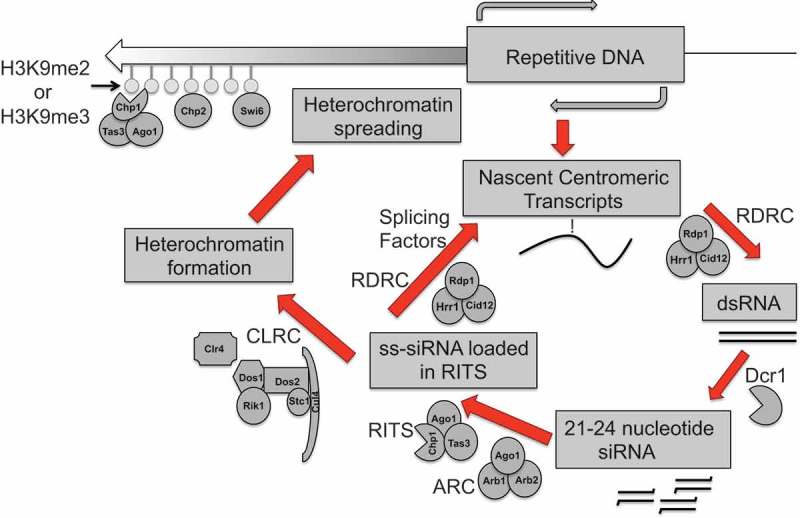
siRNA directed heterochromatin formation in fission yeast. This figure is adapted from [45,203,204] Rdp1, a member of the RDRC complex converts nascent transcripts from repetitive regions of the genome during S phase of the cell cycle into double-stranded RNA. The RNase III–like enzyme Dcr1, can process these double-stranded RNAs into siRNAs. These siRNAs then pass through the first Argonaute chaperone complex, ARC prior to loading into the RITS complex, where Ago1 endonuclease activity cleaves the passenger strand siRNA forming an effector complex that is recruited to chromatin by siRNA-nascent RNA base-pairing. This is followed by the recruitment of the CLRC histone-modifier complex to chromatin to reinstate transcriptional gene silencing by Clr4-mediated H3K9 methylation. The chromodomain protein Swi6 that along with Clr4 recruits other chromodomain proteins such as Chp1 and Chp2 then recognizes the H3K9 mark. This complex then methylates the adjacent nucleosome thus ‘spreading’ the H3K9 heterochromatin mark in an RNA interference-independent manner. The RITS effector complex can also target complementary nascent transcripts and recruit RDRC to promote the synthesis of double-stranded RNA.
