Figure 4.
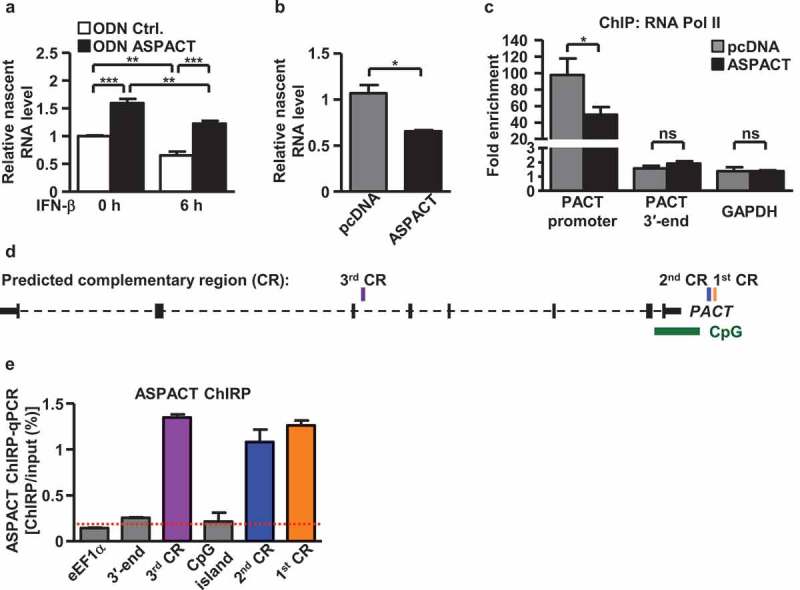
ASPACT negatively regulates PACT expression at the transcriptional level. a, b Nuclear run-on assay was performed to measure levels of nascent PACT RNA in ASPACT knockdown and IFN-treated (a) or ASPACT overexpressing (b) HeLa cells, followed by qPCR analysis. Prior to fold-change calculations, target gene expression levels were normalized to that of EF1A. The mock treatment represents as the reference. c ChIP assay was performed to demonstrate RNA Pol II occupancy of the PACT promoter chromatin. Precipitated DNA fragments were analysed by qPCR and normalized to the value of IgG. The extent of occupancy was compared between control and ASPACT-overexpressing HeLa cells. d Predicted complementary region (CR) in the PACT gene locus with potential correspondence to the ASPACT sequence. Three distinct CRs were uncovered based on high sequence alignment scores, and are represented as rectangles in relation to the CpG island and the gene body of PACT. e ChIRP analysis of ASPACT binding to the PACT chromatin. Retrieval of PACT chromatin DNA was validated by qPCR analysis of the indicated regions (using EF1A as the control). Results are expressed as retrieval rate of chromatin DNA relative to the input. For the bar graphs shown in this figure, data represent the means and SEM (error bars) from four independent experiments. Significant differences were verified by Student’s t test (ns: P > 0.05, *: P < 0.05, **: P < 0.01, ***: P < 0.001).
