Figure 6.
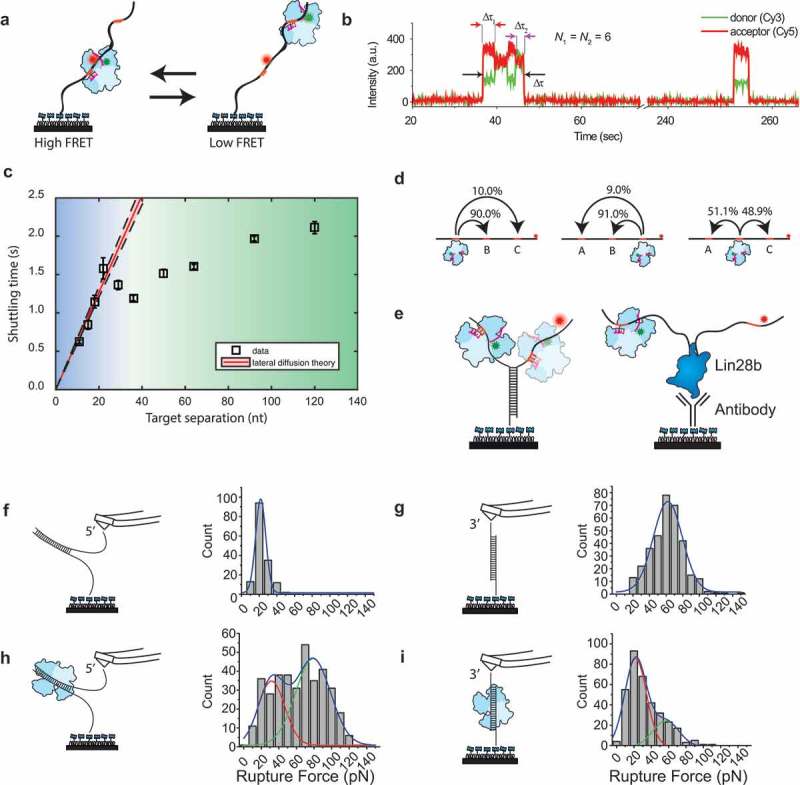
Single-molecule experiments on ago target search.
(a) Tandem target assay used by Chandradoss et al. [67]. The RNA target contains two identical targets, where the Cy5 dye is placed closer to the bottom target. During the experiments, hAgo2 shuttles between the two target sites. (b) A typical shuttling trace for N = 6 (nt 2–7). Binding to one target brings the donor dye of the guide closer to the acceptor dye, resulting in a higher FRET value. This way, binding to one target can be readily distinguished from binding to the distal target [67]. (c) Shuttling time of CbAgo plotted versus the distance between two identical targets. The red line indicates a lateral diffusion fit derived from a minimal kinetic model. The data diverge from the kinetic theory beyond the blue region and follows a different trend in the green region [72]. (d) Triple target assay used to visualize whether CbAgo skips over the middle target when translocating from site A to site C. Percentages indicate the relative amount of transitions from an initial state to a final state versus the total number of transitions [72]. (e) Left: a Y-fork construct that prevents CbAgo to slide from the target on one strand to the other. Right: Lin28 is immobilized as a protein blockade [72]. (f) DNA unzipping with AFM in absence of TtAgo. The AFM was retracted with a velocity of 200 nm/s. The rupture force is plotted with a Gaussian probability distribution fitted on top of it (blue) centred at 21.7 pN [75]. (g) DNA shearing with AFM in absence of TtAgo, with a fit centred at 62.0 pN [75]. (h) DNA unzipping with AFM in presence of TtAgo, fitted with a peak centred at 31.7 pN and 78.9 pN [75]. (i) DNA shearing with AFM in presence of TtAgo fitted with a peak centred at 21.9 pN and 56.8 pN [75]. Permission has been obtained for the above figures. Copyright 2018 American Chemical Society.
