Figure 1.
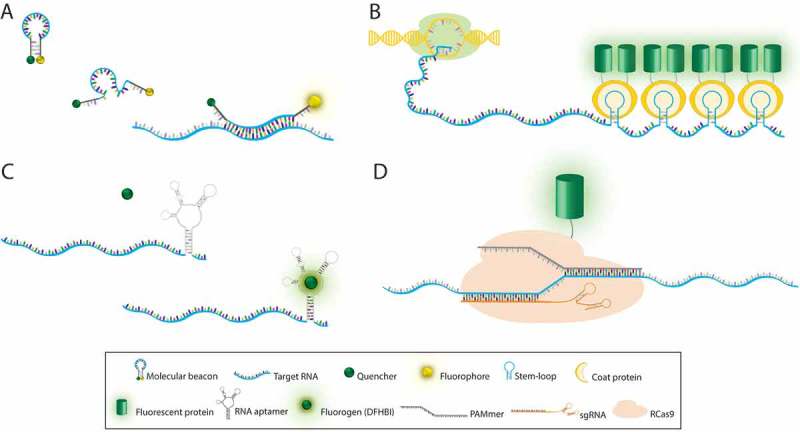
Labeling methods for mRNA imaging in vivo. (A) Molecular beacon is a single-stranded probe with hairpin-loop conformation containing a quencher and a fluorophore at 3´ and 5´ ends, respectively. Hybridization of the loop sequence to its complementary RNA sequence leads to emission of fluorescence. (B) In RNA stem-loop based labeling systems, multiple repeats of a stem-loop sequence are added to the gene of interest. Following transcription, coat proteins fused with a fluorescent protein bind to the stem-loops in the target RNA. (C) RNA aptamer probes are typically inserted into the 3′UTR of the target RNA. Binding of fluorogen (DFHBI) to the RNA aptamer triggers fluorescence emission. (D) RNA-targeting Cas9 (RCas9) system includes sgRNA and PAMmer that are both complementary to the target RNA. Nuclease-inactive Cas9 fused to a fluorescence protein enables tracking of endogenous RNA. RCas9/target RNA complex is formed in the nucleus and then exported to the cytoplasm.
