Figure 1.
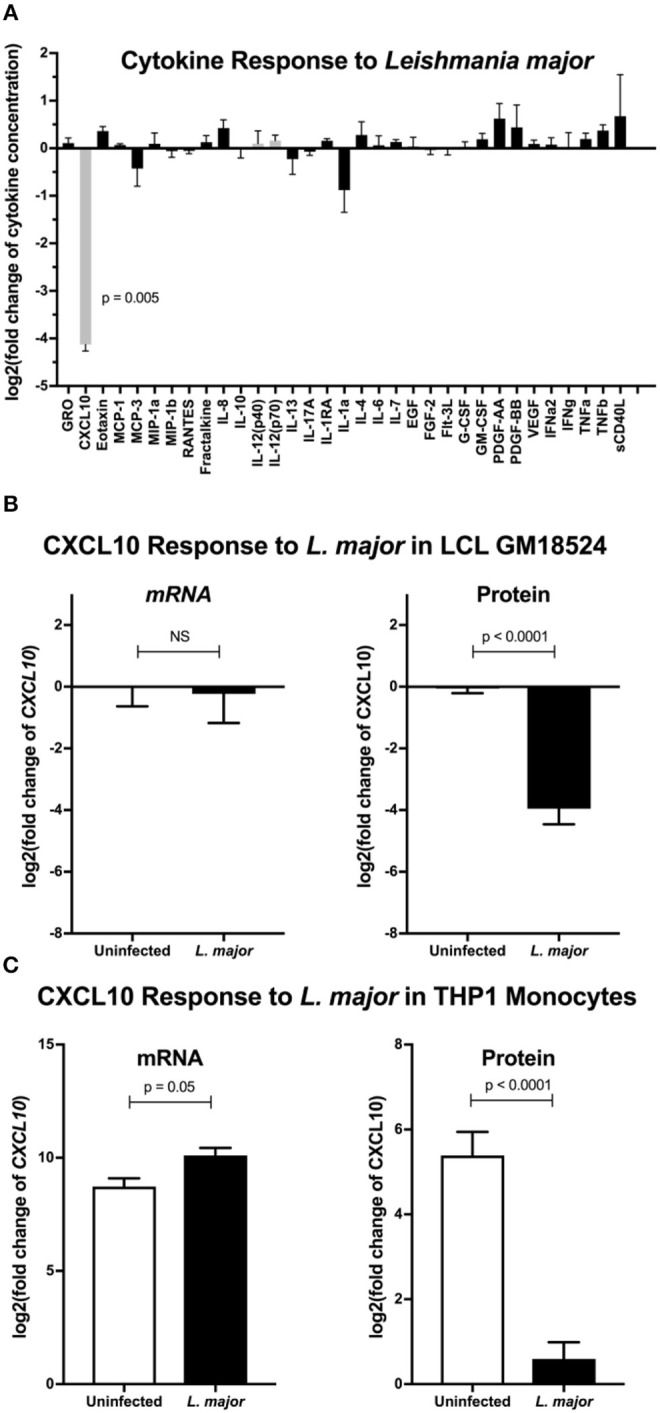
Leishmania major suppresses CXCL10 post-transcriptionally in multiple human cell lines. (A) Cytokine screening of LCLs exposed to L. major demonstrated suppression of CXCL10. Three lymphoblastoid cell lines (LCL), GM07357, GM18524, and GM19203, were infected with L. major. Chemokines secreted into culture supernatants were analyzed by Luminex. Cytokines below the limit of detection were removed from the final analysis. Values are represented as log2 of the fold change relative to uninfected LCLs. Type-1 associated cytokines are represented in gray. P-value represents Dunnett's post-hoc test compared to 1, after repeated measures one-way ANOVA. (B) CXCL10 suppression by L. major is transcriptionally independent in LCL GM18524. LCL GM18524 was infected with L. major at MOI 10 to confirm the CXCL10 suppression phenotype. Despite significant reduction in CXCL10 protein, there was no change in relative CXCL10 mRNA. CXCL10 mRNA was measured by qRT-PCR TaqMan assay using the ΔΔCt method with 18s as housekeeping gene, and CXCL10 protein was measured by ELISA. Four experimental replicates were used to calculate mRNA (n = 4) and ELISA (n = 8) fold change relative to uninfected LCL 18524. P-values calculated by Student's t-test. (C) CXCL10 produced by LPS stimulated THP-1 monocytes was suppressed by L. major. THP-1 monocytes were stimulated with LPS prior to L. major infection. Three experimental replicates were used to calculate mRNA (n = 3) and protein (n = 6) fold change relative to unstimulated, uninfected THP-1s. P-values calculated by Student's t-test.
