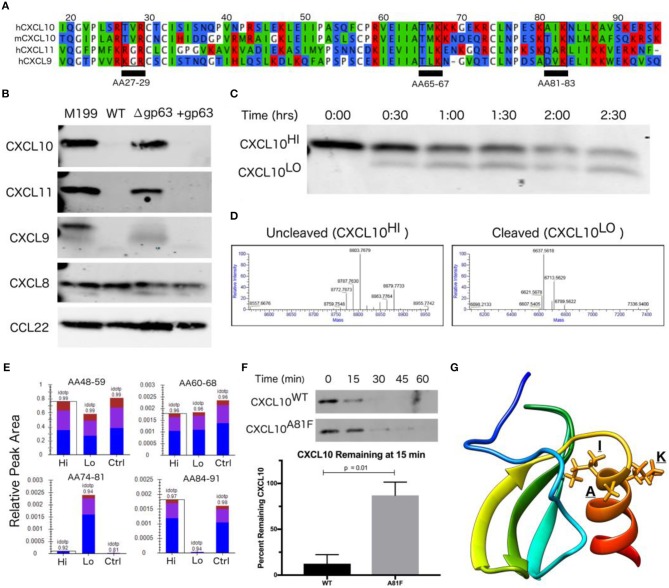
Figure 3.
CXCL10 cleavage by GP63 occurs between positions A81 and I82. (A) CXCL9/10/11 share significant homology at the amino-acid level. Multisequence alignment demonstrates that physical characteristics of amino acids are conserved across the CXCL10 family of chemokines. There are three putative GP63 cleavage sites (underlined) based on the consensus sequence of polar (P1), hydrophobic (P1′), basic (P2′) (Bouvier et al., 1990). (B) GP63 selectively cleaves chemokine ligands of the CXCR3 receptor. Conditioned media from L. major WT, Δgp63, and Δgp63+1 was incubated with human recombinant chemokines for 12 h and product detected by western blot. Cleavage is only detected for the CXCL9/10/11 family. Representative blots are shown from at least two independent experiments. (C) Cleavage by GP63 generates a smaller molecular weight protein. A time course of cleavage of human CXCL10 by heterologously expressed GP63 demonstrated an intermediate cleavage product, resolved by PAGE and Coomassie staining. (D) Cleavage by GP63 results in a change in CXCL10 molecular weight of 2.2 kD. Capillary electrophoresis-Mass Spectrometry (CE-MS) determined the molecular weight of the uncleaved (CXCL10Hi) and cleaved (CXCL10Lo) bands as 8.8 and 6.6 kD, respectively. (E) Comparative analysis by trypsin digest of cleaved and uncleaved CXCL10 reveals cleavage occurring between A81-I82. Liquid chromatography-mass spectrometry (LC-MS) following trypsin digest of CXCL10Hi and CXCL10Lo identified peptide ending at A81, exclusively in the CXCL10Lo band, and a corresponding lack of peptide coverage from AA84-91. (F) Mutation of A81F significantly impairs GP63 cleavage of CXCL10. In the presence of GP63, CXCL10A81F (n = 4 from 4 experiments) remains stable for up to 45 min whereas CXCL10WT (n = 3 from 3 experiments) degradation is nearly complete by15 min. Percentage of GP63 remaining at 15min is plotted. P-value calculated by Student's t-test. (G) The GP63 cleavage site is found on the C-terminal alpha-helix loop of CXCL10. Based on the NMR crystal structure of CXCL10 (Booth et al., 2002), the A81, I82, K83 (P1, P1′, P2′) GP63 cleavage motif maps to an exposed alpha-helical region.