Figure 3.
Protein Complex Membership for Predicting the Function of Malaria Uncharacterized Proteins
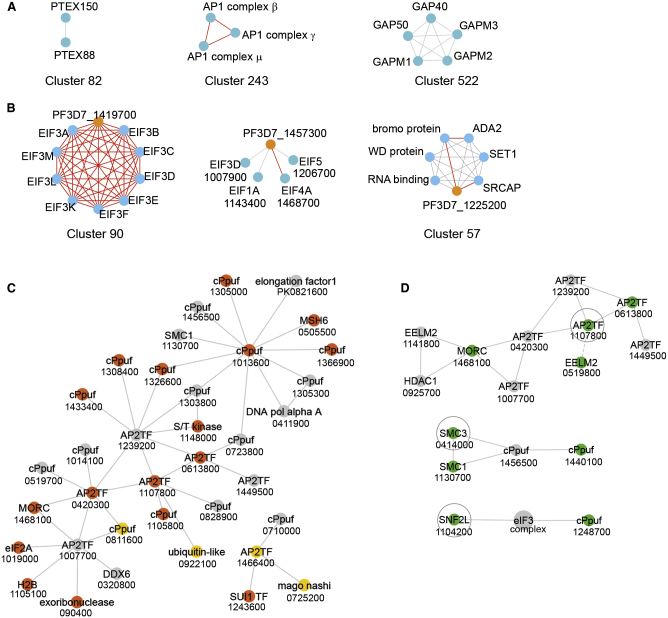
(A) Examples of clusters representing known malaria-specific protein complexes PTEX (de Koning-Ward et al., 2009), AP-1 (Kaderi Kibria et al., 2015), and glideosome (Frénal et al., 2017) found in this study.
(B) Examples of clusters containing conserved Plasmodium proteins of unknown function (in orange). Red edges represent interactions annotated in STRING.
(C) The ApiAP2 transcription factor interaction network. Relevant first-order interactions involving ApiAP2 transcription factors were extracted from the PPI network. Nodes labeled cPpuf are conserved Plasmodium proteins of unknown function. Orange nodes are essential proteins, and yellow nodes represent proteins whose mutation results in slow growth.
(D) Validation of interactions by affinity purification-mass spectrometry from tagged P. berghei lines. Represented are subsections of the PPI network. The baits are surrounded by a circle. The proteins identified specifically in the immunoprecipitate of each bait are in green. The data are from two independent biological repeats. The dotted line represents an indirect link in the network.
The numeric part of P. falciparum gene names is shown in (B)–(D).
See also Table S8.