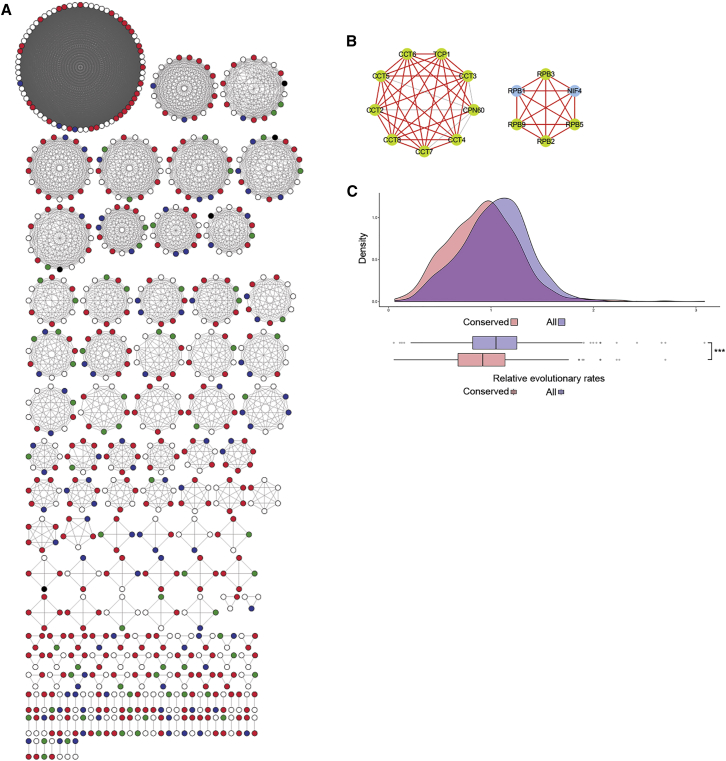
Figure 4.
A Conserved Malaria Interaction Network
(A) Clusters were generated from the protein interactions common to all three Plasmodium species studied, visualized as network graphs. The essential proteins are shown as red nodes, the blue nodes are proteins whose absence leads to slow growth, and the green nodes are proteins that cause no growth phenotype when absent.
(B) Examples of conserved clusters detected in all three Plasmodium species. Interactions between the green nodes were detected in all of the species; the interactions with the blue nodes were not detected in P. knowlesi. The red edges represent the interactions annotated in STRING.
(C) Density distribution and boxplots of relative evolutionary rates of OrthoMCL protein families to P. falciparum. The plots for the high-confidence PPI network and of protein families in the clusters from (A) are shown. The differences in distribution were assessed with a Wilcoxon rank-sum test (∗∗∗p < 2.9e−14).
See also Table S9.