Figure 1.
Disruption of 53BP1 Higher Order Oligomerization Abrogates CSR without Affecting DSB End Protection
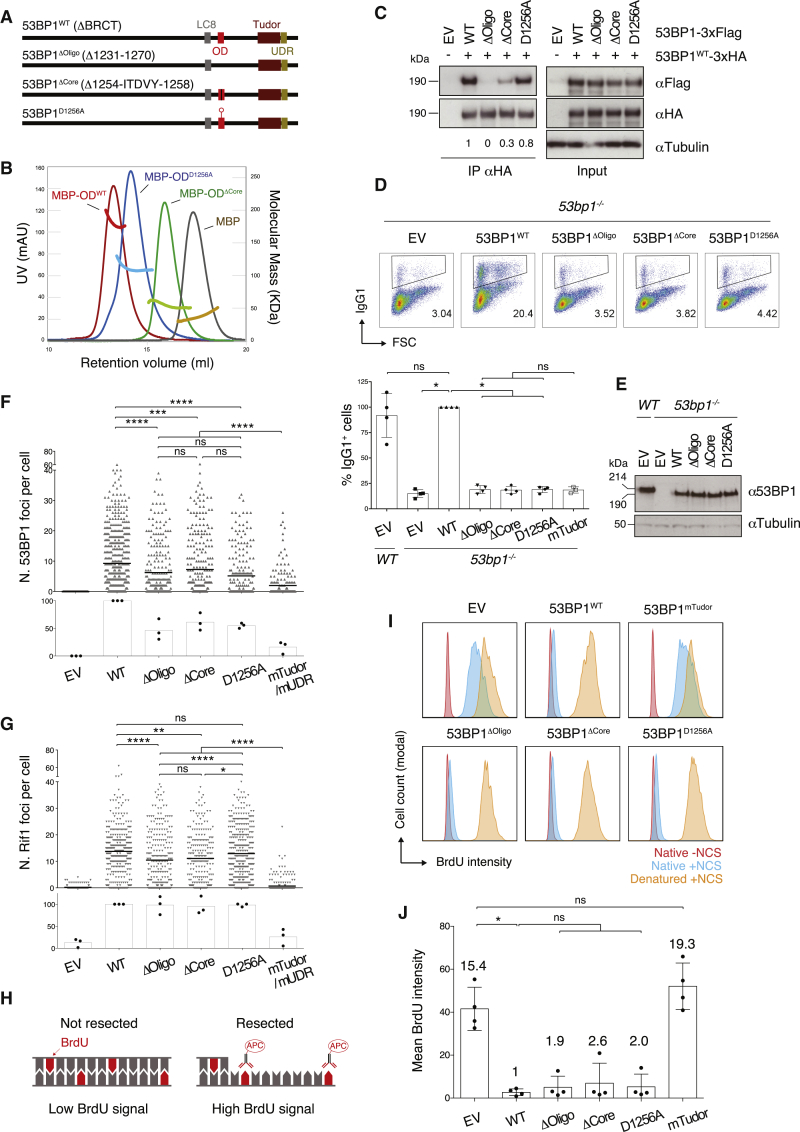
(A) Schematic representation of 53BP1 protein domains and OD mutants. LC8, LC8 (8 kDa light chain dynein)-binding motif; OD, oligomerization domain; Tudor, Tudor domain; UDR, ubiquitylation-dependent recruitment motif; ΔBRCT, BRCA1 C terminus domains-truncated.
(B) Gel filtration coupled to right-angle light scattering (RALS) analysis of the OD constructs fused to N-terminal MBP. UV absorbance is shown in red-blue-green-brown and refers to the left y axis, whereas the molecular mass of the constructs as deduced by RALS is shown in the corresponding brighter colors and refers to the right y axis. Results are representative of two independent experiments. See also Figure S1.
(C) Western blot (WB) analysis of anti-HA immunoprecipitates from irradiated (20 Gy; 1 h recovery time) BOSC23 cells transfected with the indicated 53BP1-3xFlag/-3xHA constructs. Numbers underneath each lane indicate relative quantification of oligomutant versus WT 53BP1-3xFlag proteins in anti-HA immunoprecipitates (53BP1WT-3xFlag set to 1). Results are representative of two independent experiments. EV, empty vector.
(D) Top: representative flow cytometry plots measuring CSR to IgG1 in WT and 53bp1−/− splenocytes transduced with the indicated constructs. Numbers in the plots refer to the percentage of switched cells (IgG1+ cells). Bottom: summary dot plot for four independent experiments. CSR efficiencies within each experiment were normalized to the CSR value of 53BP1WT-reconstituted cells, which was set to 100%.
(E) Representative WB analysis of 53bp1−/− B lymphocytes reconstituted with the indicated 53BP1 viral constructs.
(F and G) Automated IRIF quantification of immunofluorescent staining for 53BP1 (F) and Rif1 (G) in irradiated (10 Gy; 2 h recovery time) 53bp1−/− iMEFs reconstituted with the indicated constructs. The top graph within each panel shows a representative experiment, and the bottom graph summarizes the mean values of three experiments performed on three independent sets of reconstituted cell lines. Each symbol in the top graph represents a single cell, and the number of scored cells was ≥200 per sample. The mean is indicated. Either 53BP1mTudor or 53BP1mUDR was included in parallel to EV as an additional control in the individual repeats. The representative experiments in (F) and (G), top graphs, used 53BP1mTudor. See also Figure S2B for representative immunofluorescent staining images.
(H) Schematic representation of the BrdU assay for DNA end resection. BrdU treated cells were analyzed for BrdU content under native (not denatured) conditions to estimate resection levels following NCS treatment. Adapted from (Tkáč et al., 2016).
(I) Representative BrdU fluorescence-activated cell sorting (FACS) plots showing levels of resection in NCS-treated 53bp1−/− iMEF reconstituted with the indicated constructs.
(J) Summary graph showing quantification of the BrdU FACS data represented in (I) for four experiments performed on three independent sets of reconstituted cell lines. Numbers above each bar indicate the fold mean BrdU intensity over WT, which was set to 1.
Significance in (D), (F), (G), and (J) was calculated using the Mann-Whitney U test, and error bars represent SD. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ∗∗∗∗p ≤ 0.0001; ns, not significant. See also Figure S3.