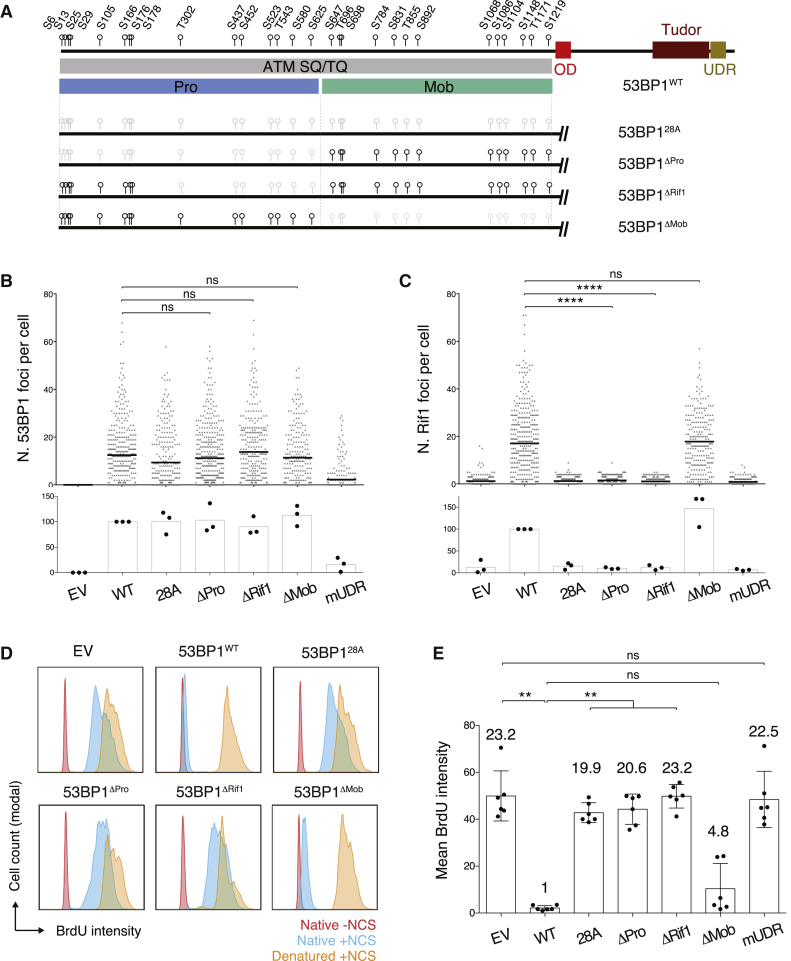
Figure 2.
Inactivation of 53BP1 SQ/TQ Sites T302 to S625 Is Sufficient to Abolish Rif1-Dependent Inhibition of DNA End Resection
(A) Top: schematic representation of 53BP1 protein with the position of the 28 N-terminal SQ/TQ motifs indicated as black circles. Bottom: schematic representation of the 53BP1 phospho mutants with S/TQ sites indicated as black circles and S/TQ-to-AQ substitutions marked in light gray. Pro, DNA end protection SQ/TQ set; Mob, DNA mobility SQ/TQ set.
(B and C) Automated IRIF quantification of immunofluorescent staining for 53BP1 (B) and Rif1 (C) in irradiated (10 Gy; 2 h recovery time) 53bp1−/− iMEFs reconstituted with the indicated constructs. The top graph within each panel shows a representative experiment, and the bottom graph summarizes the mean values of three experiments performed on three independent sets of reconstituted cell lines. Each symbol in the top graph represents a single cell, and the number of scored cells was ≥200 per sample. The mean is indicated. See also Figure S2D for representative immunofluorescent staining images.
(D) Representative BrdU FACS plots showing levels of resection in NCS-treated 53bp1−/− iMEFs reconstituted with the indicated constructs.
(E) Summary graph showing quantification of the BrdU FACS data represented in (D) for six experiments performed on three independent sets of reconstituted cell lines. Numbers above each bar indicate the fold mean BrdU intensity over WT, which was set to 1.
Significance in (B), (C), and (E) was calculated using the Mann-Whitney U test, and error bars represent SD. ∗∗p ≤ 0.01 and ∗∗∗∗p ≤ 0.0001; ns, not significant. See also Figure S4B.