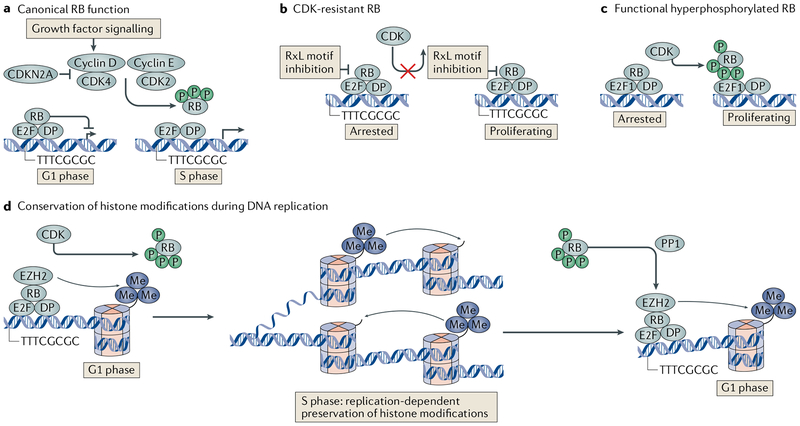
Fig. 1 |. Canonical RB-E2F regulation and mechanisms of CDK resistance.
a | An illustration of our definition of canonical RB function is presented. In this model, growth factors signal the expression and activation of D-type cyclins. Cyclin D-cyclin-dependent kinase 4 (CDK4) or CDK6 and cyclin E-CDK2 act to hyperphosphorylate RB. Members of the E2F family of transcription factors and their dimeric partners (DPs) are released to activate the expression of genes that advance the cell cycle. This signalling pathway can be blocked by CDK inhibitor 2A (CDKN2A)-mediated inhibition of cyclin D, which is associated with CDK activity, b | RB can escape CDK hyperphosphorylation when its RxL motif is bound by protein phosphatase 1 (PP1) or when the RxL motif is acetylated or methylated, collectively depicted as RxL motif inhibition. This prevents substrate recognition and ensures retention of RB function. c | RB and transcription factor E2F1 can interact through molecular contacts that are distinct from RB and other E2F family members. In this interaction, CDK hyperphosphorylation of RB is unable to disrupt E2F1 binding. Similarly, ultraviolet light or osmotic shock can activate p38 MAPK to phosphorylate RB. Once phosphorylated, this stimulates E2F1 binding, even when RB is hyperphosphorylated by CDKs. d | RB-E2F complexes can recruit chromatin-modifying enzymes such as enhancer of zeste homologue 2 (EZH2) to mediate patterns of methylation of histone H3 lysine 27 (H3K27me3) at promoters. RB is removed from these locations by CDK hyperphosphorylation at the onset of S phase, but histone tail modification patterns are preserved during DNA replication, enabling stable transmission of the epigenetic state to the next cell generation. In the ensuing G1 phase, RB is dephosphorylated by PP1A and RB-E2F recruitment of histone methyltransferase activity reinforces patterns of modification in the ensuing cell cycle.