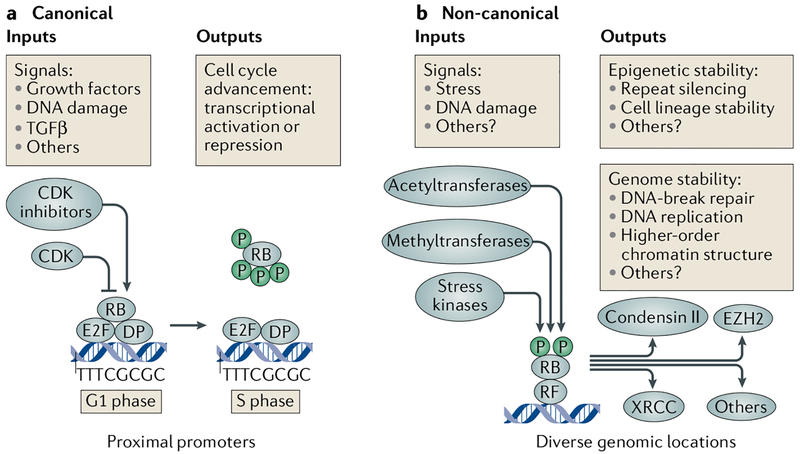
Fig. 4 |. Features of the canonical and non-canonical RB pathways.
a | Positive growth signals activate cyclin-dependent kinases (CDKs) to phosphorylate and inactivate RB, whereas negative growth regulators (DNA damage and transforming growth factor-β (TGFβ)) augment CDK inhibitors. Inactivation of RB in the canonical pathway leads to E2F target gene transcription. b | Non-canonical RB functions are best defined by their ambivalence to CDK regulation. Signals that likely activate non-canonical RB include stresses that activate p38 MAPK phosphorylation and DNA damage that stimulates P300-associated factor (PCAF)-dependent acetylation and SET domain-containing protein 8 (SET8; also known as KMT5A) or SET and MYND domain-containing protein (SMYD)-dependent methylation. With the aid of recruitment factors (RFs), often transcription factor E2F1, RB is capable of maintaining genome stability by localizing to sites of DNA breaks and stimulating non-homologous end joining or homologous recombination repair in complex with the DNA binding proteins X-ray repair cross-complementing protein 5 (XRCC5), XRCC6 or transcription activator BRG1. RB also ensures fidelity of DNA replication and chromosome condensation through direct interactions with condensin II complexes and the indirect influence of cohesin recruitment. Lastly, RB recruits enhancer of zeste homologue 3 (EZH2) to trimethylate histone H3 lysine 27 (H3K27me3) and inhibit enhancers to control lineage commitment and to silence expression of repetitive sequences. Importantly, this diverse grouping of functions is facilitated by the association of RB with the genome over a much larger scale than proximal promoters. DP, dimeric partner.