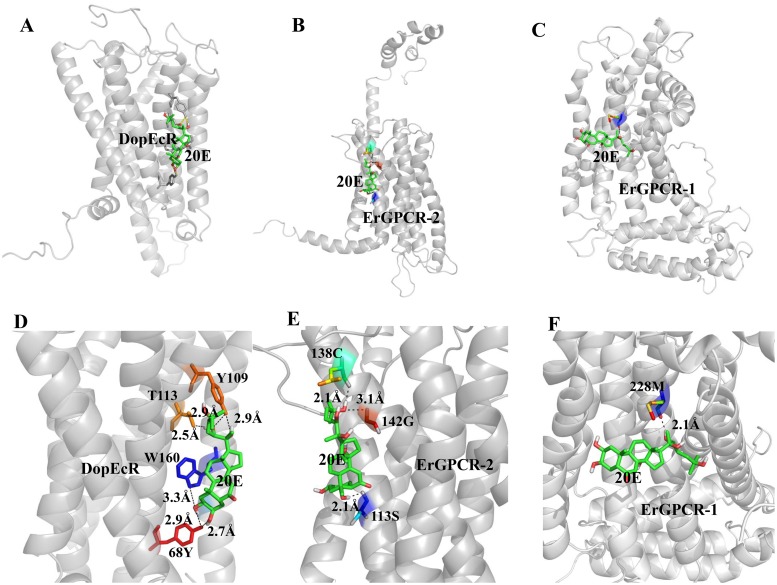
Fig 8. Modeling of the ligand-binding complex of the GPCRs.
A. B. and C. Prediction of the Surflex-Dock (SFXC) program from the SYBYL X2.0 software. Overall structures of DopEcR, ErGPCR-2, and ErGPCR-1 (gray) and docked 20E (Green), respectively. D. E. and F. A closer view of the pockets relative to docking models of DopEcR-, ErGPCR-2- and ErGPCR-1-20E complex and the amino acid residues mutated in this study. The dotted lines indicate the predicted hydrogen bonds between the amino acid residues of GPCRs and 20E. Orientation is the same in all models.