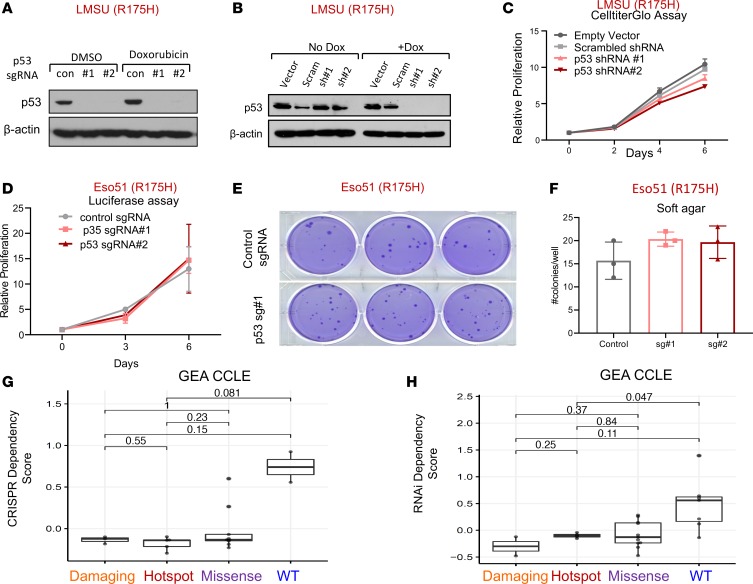
Figure 2. Mutant p53 is not required for primary tumor functions of gastroesophageal cancer cells.
(A) Immunoblot showing protein expression levels of p53 in the LMSU gastric adenocarcinoma cell line, which harbors an endogenous R175H p53 mutation, stably expressing a Cas9 control (con) vector or 2 distinct targeting sgRNAs in addition to Cas9. (B) Immunoblot showing protein expression levels of p53 in LMSU gastric cancer cells expressing an inducible vector control, scrambled (Scram) control, or 2 targeting shRNAs with or without doxycycline (Dox). (C) Proliferation of LMSU gastric adenocarcinoma cells expressing a constitutively active vector control, scrambled control, or 2 p53-targeting shRNAs using CellTiter-Glo. (D) Proliferation of Eso51 nonadherent esophageal cancer cells (R175H) stably expressing firefly luciferase in addition to a constitutively active Cas9 control vector or 2 p53-targeting sgRNAs using a luciferase assay. (E) Soft agar colony formation assay of Eso51 esophageal adenocarcinoma cells (R175H) stably expressing a constitutively active Cas9 control vector or 1 p53-targeting sgRNA. (F) Quantification of soft agar colony formation assay shown in E. (G and H) TP53 dependency scores for GEA cell lines derived from the CCLE, with damaging (nonsense/frameshift) mutations, hotspot (R175H, R248Q/W, R273C/H) mutations, or missense mutations or WTTP53 using (G) CRISPR and (H) RNAi dependency data. Lines within boxes represent median, the bounds of the boxes represent the 25th and 75th percentiles, and the whiskers extend to the lowest/largest values within 1.5 IQR from the lower and upper quartiles, respectively. P values were calculated by Wilcoxon’s rank-sum test