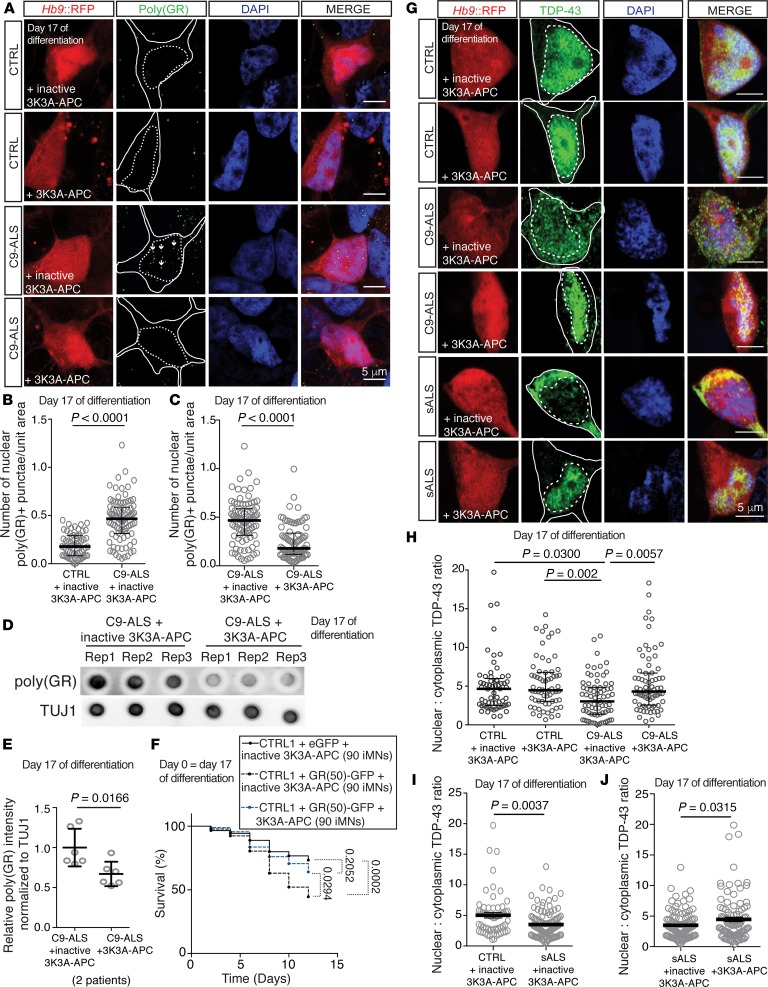
Figure 3. Rescue of autophagosome formation by 3K3A-APC improves proteostasis.
(A–C) Immunostaining (A) and quantification (B and C) to determine endogenous poly(GR)+ punctae in control or C9-ALS iMNs with 10 nM inactive 3K3A-APC or 3K3A-APC treatment for 6 days. Quantified values represent the average number of nuclear poly(GR)+ punctae in n = 30 iMNs (controls) or 40 to 44 iMNs (C9-ALS) per line per condition from 2 control or 2 C9-ALS patient lines. For each line, iMNs were quantified from 2 independent iMN conversions per line per condition. Median ± interquartile range. Each gray circle represents the number of poly(GR)+ punctae/unit area in a single iMN. Mann-Whitney testing. Solid and dotted lines in A outline the cell body and nucleus, respectively. Scale bars: 5 μm. (D and E) Dot blot (D) and quantification (E) of poly(GR)+ levels in iMNs from 2 C9-ALS patient lines with 10 nM inactive 3K3A-APC or 3K3A-APC treatment for 6 days. Each gray circle represents 1 dot blot sample. Mean ± SD. n = 3 independent iMN conversions per line per condition. One-way ANOVA with Tukey’s correction across all comparisons. (F) Survival of control iMNs without excess glutamate with overexpression of eGFP or GR(50)-eGFP and 10 nM inactive or active 3K3A-APC. n = 90 iMNs per condition, iMNs quantified from 3 biologically independent iMN conversions. Two-sided log-rank test, corrected for multiple comparisons, statistical significance was calculated using the entire survival time course. n = 90 iMNs per condition. (G–J) Immunofluorescence analysis of total TDP-43 (G) and quantification of the ratio of nuclear to cytoplasmic TDP-43 in control, C9-ALS (H), or sporadic ALS iMNs (I and J). Ratio of nuclear to cytoplasmic TDP-43 in individual C9-ALS iMNs treated with 10 nM inactive 3K3A-APC or 3K3A-APC for 6 days. iMNs from 2 controls, 2 C9-ALS, and 4 sporadic ALS patients were quantified. n = 30 iMNs per line (control and C9-ALS) per condition or n = 26 iMNs (I), 30 iMNs (J) (inactive 3K3A-APC), or n = 35 iMNs (J) (3K3A-APC) (sporadic ALS) per condition per line from 2 biologically independent iMN conversions were quantified. Each gray circle represents a single iMN. For H, median ± interquartile range. Kruskal-Wallis testing. For I and J, mean ± SEM. Unpaired t test with Welch’s correction. Scale bars: 5 μm. Dotted lines outline the nucleus and cell body. The day of differentiation stated on each panel indicates the day of differentiation on which the experimental treatment or time course was initiated.