Abstract
Background:
Mother-to-newborn transmission of obesity-associated microbiota may be modified by delivery mode. Prospective data to test this hypothesis are still sparse.
Objective:
To examine prospective associations of maternal pre-pregnancy BMI and gestational weight gain with the infant gut microbiome by delivery-mode strata.
Subjects/Methods:
In 335 mother-infant pairs in the New Hampshire Birth Cohort, we ascertained data from questionnaires and medical records, and generated microbiome data from 6-week-old infants’ stool using Illumina 16S rRNA gene sequencing (V4-V5 region). Analyses were stratified by delivery mode and conducted before and after adjusting for potential confounders, which included maternal age, education, parity, and Mediterranean diet score.
Results:
Among 335 mothers, 56% had normal pre-pregnancy BMI (<25, referent), 27% were overweight (BMI 25.1–30), and 18% obese (BMI >30). Among the 312 mothers with weight gain data, 10% had inadequate weight gain, 30% adequate (referent), and 60% excess. In the vaginal-delivery group, maternal overweight or obesity was associated with higher infant gut microbiome diversity and higher abundance of 15 operational taxonomic units (OTUs), including overrepresentation of Bacteroides fragilis, Escherichia coli, Veillonella dispar and OTUs in the genera Staphylococcus and Enterococcus. Delivery mode modified associations of pre-pregnancy BMI with several genera, including the most abundant genus, Bacteroides (P for interaction = 0.05). In the Cesarean-delivered group, there were no significant associations between pre-pregnancy BMI, or gestational weight gain categories with infant microbiome (alpha) diversity or OTUs.
Conclusions:
Among vaginally-delivered infants, maternal overweight and obesity was associated with altered infant gut microbiome composition and higher diversity. These associations were not observed in Cesarean-delivered infants, whose microbiome development, we have shown, differs from vaginally-delivered infants. Our study provides additional evidence of delivery-mode dependent associations of maternal weight status with the infant gut microbiota. The role of these associations in mediating the intergenerational cycle of obesity warrants further examination.
Introduction
Overweight and obesity (OWOB), particularly during childhood, is a major public health concern due to its continued rise in prevalence in the United States (1, 2) and throughout the world (3). Among children under 5, the global prevalence of OWOB rose from 30 million in 2000 to 41 million by 2016, with an additional 240 million OWOB children aged 5 – 19 years in 2016 (4, 5). The global rise in OWOB, along with the intractable challenge of achieving sustained weight loss in OWOB individuals later in life, has led to an increased interest in the early-life origins and primordial prevention of OWOB.
An important area of research on the primordial prevention of OWOB aims to elucidate, and eventually intervene upon, the mechanisms underpinning the intergenerational cycle of obesity between a mother and her child. Mothers that are OWOB are two to four times more likely to give birth to children who will become OWOB, starting as early as 2 to 5 years of age (6–8). Yet genome wide association studies have found that genetic polymorphisms likely only explain a small fraction of this phenomenon (9). One potential, yet understudied pathway that may help explain the intergenerational cycle involves the mother-to-newborn transmission of microbiota.
Because the newborn gut microbiota is seeded at birth (10, 11), it has been postulated that differential mother-to-newborn transfer of microbiota may perpetuate the intergenerational cycle of obesity (12). In our review on this topic, we concluded that there is a dearth of knowledge with respect to the effects of maternal pre-pregnancy BMI and gestational weight gain on the infant microbiota, with conflicting results from the few existing studies (13). Since our review, Tun et al. reported delivery-mode specific associations between maternal pre-pregnancy BMI and the infant gut microbiome at 3–4 months (14). This finding is consistent with our previous investigation on this topic (15), further supporting the hypothesis that birth mode may modify the association between maternal weight and infant microbiome development.-
In the current study we aimed to extend previous findings on this topic by rigorously examining associations of pre-pregnancy BMI and gestational weight gain with the 6-week-old infant gut microbiome in the New Hampshire Birth Cohort. We conducted analyses stratified by delivery mode because it has been implicated in modifying associations between maternal weight status and the infant gut microbiome (14–16). Consistent with the notion of differential mother-to-newborn vertical transmission of bacteria at birth, in particular Bacteroides spp. (10–12), and with our prior observations (15), we hypothesized that in vaginally-delivered infants maternal pre-pregnancy overweight and obesity (vs. normal weight) would be associated with altered infant gut microbiota diversity and composition.
Methods
Study Population
Detailed information on the New Hampshire Birth Cohort has been described previously (16). Briefly, in 2009, women who were 18 to 45 years of age and between 24 and 28 weeks of gestation were recruited for participation in an observational birth cohort study. Mothers and their infants were then followed up at 6 weeks post-delivery. This timing was chosen due to its alignment with an established 6-week post-partum visit, and because infant feeding practices (breastfeeding v. formula feeding) are often well established by this age. During this visit, infant stools were collected for microbial community profiling. Data on covariates of interest were also collected using a combination of medical records, questionnaires, and telephone-calls.
Pre-pregnancy BMI was derived from self-reported weight and height from a prenatal questionnaire. Gestational weight gain information was ascertained from a postpartum questionnaire. We excluded 2 women missing pre-pregnancy BMI data, and 8 women with pre-pregnancy BMI < 18.5 kg/m2 (flowchart displayed in Figure S1). We then categorized the remaining 335 women as normal weight (18.5 kg/m2 ≤ BMI < 25 kg/m2), overweight (25 kg/m2 ≤ BMI < 30 kg/m2), or obese (BMI ≥ 30 kg/m2). For gestational weight gain, we categorized the 312 women who also had gestational weight gain data as having experienced inadequate, adequate, or excess weight gain based on Gilmore and Redman’s method (17), which relies on the Institute of Medicine’s recommended rates of weight gain by pre-pregnancy BMI category (18). The recommendations state that during the second and third trimesters of pregnancy, normal weight mothers are expected to gain 0.8 – 1 lb/wk, overweight mothers 0.5 – 0.7 lb/wk, and obese mothers 0.4 – 0.6 lb/week.
Microbial Sampling and 16s rRNA Sequencing
Infant stools were collected from diapers and aliquoted within 24 hours into sterile tubes and stored at −80°C. DNA extraction was then conducted using Zymo DNA extraction kits (Tustin, CA, USA) on thawed samples, followed by quantity and purity assessment using OD260/280 nanodrop measurement (19). Fusion primers were used to obtain 16S ribosomal RNA (rRNA) V4-V5 amplicons. One of eight five-nucleotide long forward primers was used to connect the Illumina-specific bridge and sequencing primer regions with the 16S region, whereas the reverse primer was simply one of 12 Illumina indices.
16S rRNA amplification of the V4-V5 hypervariable regions was conducted at the Marine Biological Laboratory in Woods Hole, Massachusetts, USA for microbial profiling using both forward and reverse primers (20, 21). Due to the use of both forward and reverse primers, 96 samples were multiplexed per lane. PCR was then conducted using triplicate samples of 33 μL, and a combination of 1.0 U Platinum Taq Hi-Fidelity Polymerase (Life Technologies Carlsbad, CA, USA), 1X Hi-Fidelity buffer, 200 μM dNTP3 PurePeak DNA polymerase mix (Pierce Nucleic Acid Technologies Milwaukee, WI, USA), 1.5 mM MgSO4 and 0.2 μM of each primer. Lastly, qPCR was used to quantify each library pool before sequencing using paired-end Illumina MiSeq 100 cycle runs.
Analytic Methods
Forward and reverse reads derived from Illumina sequencing were assembled and demultiplexed using CASAVA 1.8.2, and denoised and quality filtered using DADA2 in Quantitative Insights Into Microbial Ecology 2 (QIIME 2) version 2017.10 (22, 23). We then employed QIIME 2 to assign operational taxonomic units (OTUs) using the Greengenes database with a 99% cutoff for similarity before importing the data into R 3.4.2 using the biom-format package in Python (24–27).
Bray-Curtis and unweighted and weighted UniFrac distances were used to analyze microbial beta diversity (overall microbial community composition) after rarefying to a sampling depth of 1029 counts (28, 29). Principal coordinate analysis (PCoA) plots were created within QIIME 2 using Emperor (30). We tested for differences in beta diversity using permutational multivariate analysis of variance (PERMANOVA) in QIIME 2 with 999 permutations. We also conducted PERMANOVA tests with 10,000 permutations, after adjusting for covariates that were potential confounders, using the vegan package in R (31). We considered a p-value < 0.05 as statistically significant.
Microbial alpha diversity was estimated with observed OTUs (unique number of OTUs), Shannon diversity index, and Chao1 using the phyloseq package in R (26). Data were rarefied to the same sampling depth of 1029, with 10,000 permutations. The associations of pre-pregnancy BMI and gestational weight gain with gut microbiota alpha diversity metrics were analyzed using linear regression. We considered a p-value < 0.05 as statistically significant.
We next compared the relative abundance of the most dominant genera (those > 1.0% in prevalence) in the infants’ gut microbiota using zero-inflated beta regression in the gamlss package within R, which accounts for excessive zero counts (32). Based on previous publications (11, 15), we a priori hypothesized an association of maternal pre-pregnancy BMI categories with the Bacteroides genus in the vaginal strata. With the DESeq2 package in R, we then agnostically assessed differential relative abundance of specific OTUs using a log-transformed negative binomial model with an internal adjustment for library size (33). To account for multiple-testing in this agnostic approach, we considered a False-Discovery-Rate (FDR) adjusted p-value < 0.10 as statistically meaningful. We did not rarefy OTU counts in differential abundance analyses because we wanted to maximize use of the sequencing data available (34).
We performed all regression analyses before and after stratifying by delivery mode, our a priori hypothesized effect measure modifier, as well as before and after adjusting for potential confounders. We adjusted for covariates that were associated with both pre-pregnancy BMI or gestational weight gain and overall microbial community composition, but not on the causal pathway to avoid over adjusting our model for potential mediators. The final multivariable model included maternal age, parity, maternal educational achievement at time of pregnancy, and maternal Mediterranean diet score, excluding alcohol, during pregnancy.
We also conducted sensitivity analyses by further adjusting for maternal antibiotics, feeding method at 6 weeks (defined as exclusively breastfed vs. combination fed or exclusively formula fed) and infant sex, as well as by restricting to infants exclusively breastfed at 6 weeks.
Finally, in addition to our a priori delivery-mode specific examination of associations, we explored effect measure modification by sex through stratification. We tested the significance of these interactions by including a delivery mode—pre-pregnancy BMI interaction term in the differential abundance model for Bacteroides (the most abundant genus in our sample).
Results
Participant Characteristics
The final sample for pre-pregnancy-BMI analyses comprised 335 mother-infant pairs, and 312 pairs for gestational-weight-gain analyses. Women in our study sample had a mean pre-pregnancy BMI of 25.9 kg/m2 (SD: 5.5), with 55.5% classifying as normal weight, 26.9% as overweight, and 17.6% as obese. The mean gestational weight gain was 31.7 lbs (SD: 12.4), with 9.6% of the cohort experiencing inadequate weight gain, 27.8% adequate weight gain, and 55.8% excess weight gain. The majority of women in this cohort delivered vaginally (71.9%), exclusively breastfed their infants up to 6 weeks (69.0%), had a college degree (75.4%), and were Caucasian (99.4%).
In Table 1, we described mother-infant characteristics, jointly stratified by delivery mode and pre-pregnancy BMI categories. Among women who delivered vaginally, women with higher pre-pregnancy BMI tended to be younger, were more likely to be primiparas (delivering their first child), to have ever smoked, to have less education, and to have lower Mediterranean diet scores. Across both strata, we found that women with higher pre-pregnancy BMI had lower gestational weight gain, were less likely to exclusively breastfeed, had more male children, and were more likely to experience gestational hypertension. We show mother-infant characteristics jointly stratified by delivery mode and gestational weight gain in Table S1.
Table 1.
Mother-infant characteristics jointly stratified by delivery mode and pre-pregnancy BMI category†
| Vaginally Delivered | Cesarean Section Delivered | |||||
|---|---|---|---|---|---|---|
| Normal Weight (n = 148) | Overweight (n = 58) | Obese (n = 35) | Normal Weight (n = 38) | Overweight (n = 32) | Obese (n = 24) | |
| Maternal age, years, mean (SD) | 32.2 (4.5) | 31.2 (4.1) | 30.6 (4.3) | 33.2 (4.8) | 31.7 (4.7) | 32.9 (4.7) |
| Gestational age, wk, mean (SD) | 39.5 (1.6) | 39.88 (1.2) | 39.4 (1.9) | 39.3 (1.7) | 39.2 (2.0) | 38.9 (1.6) |
| Pre-pregnancy BMI, kg/m2, mean (SD) | 22.2 (1.6) | 26.9 (1.4) | 35.1 (4.2) | 22.3 (1.5) | 27.3 (1.3) | 36.1 (5.0) |
| Gestational weight gain, lb, mean (SD) | 33.3 (10.5) | 31.1 (12.8) | 23.8 (13.6) | 37.8 (14.8) | 30.3 (9.5) | 25.6 (12.9) |
| Exclusively breastfed at 6 weeks, n (%) | 113 (76.4%) | 40 (69.0%) | 19 (54.3%) | 28 (73.7%) | 19 (59.4%) | 12 (50.0%) |
| Infant sex, fem, n (%) | 74 (50.0%) | 32 (55.2%) | 14 (40.0%) | 28 (73.7%) | 17 (53.1%) | 12 (50.0%) |
| Infant birth weight, g (SD) | 3418.0 (513.6) | 3575.2 (414.8) | 3268.5 (513.6) | 3411.2 (556.2) | 3421.1 (476.5) | 3432.0 (591.1) |
| Parity, n (%) | ||||||
| 0 | 57 (38.5%) | 22 (37.9%) | 19 (54.3%) | 22 (57.9%) | 17 (53.1%) | 14 (58.3%) |
| 1 | 66 (44.6%) | 27 (46.6%) | 8 (22.9%) | 13 (34.2%) | 10 (31.2%) | 8 (33.3%) |
| >= 2 | 25 (16.9%) | 9 (15.5%) | 8 (22.9%) | 3 (7.9%) | 5 (15.6%) | 2 (8.3%) |
| Highest education, n (%) | ||||||
| High School or less | 12 (8.4%) | 5 (9.1%) | 4 (12.9%) | 4 (10.5%) | 2 (6.5%) | 2 (8.7%) |
| Some college | 19 (13.3%) | 14 (25.5%) | 7 (22.6%) | 5 (13.2%) | 3 (9.7%) | 2 (8.7%) |
| College degree | 51 (35.7%) | 20 (36.4%) | 9 (29.0%) | 13 (34.2%) | 16 (51.6%) | 11 (47.8%) |
| Graduate school | 61 (42.7%) | 16 (29.1%) | 11 (35.5%) | 16 (42.1%) | 10 (32.3%) | 8 (34.8%) |
| Mediterranean Diet Score, mean (SD) | 4.15 (1.70) | 3.46 (2.08) | 2.87 (1.76) | 3.92 (1.92) | 3.32 (1.49) | 3.96 (1.77) |
| Smoking, n (%) | ||||||
| Ever smoked | 126 (88.1%) | 50 (90.9%) | 27 (81.8%) | 34 (89.5%) | 29 (93.5%) | 21 (91.3%) |
| Smoked during pregnancy | 9 (6.3%) | 3 (5.5%) | 6 (18.2%) | 0 (0%) | 1 (3.2%) | 1 (4.3%) |
| Never smoked | 8 (5.6%) | 2 (3.6%) | 0 (0%) | 4 (10.5%) | 1 (3.2%) | 1 (4.3%) |
| Alcohol, drinks/wk, mean (SD) | 3.7 (5.2) | 3.1 (4.5) | 2.3 (4.4) | 3.5 (2.9) | 5.4 (5.8) | 2.0 (3.2) |
| Prenatal antibiotics, n (%) | 22 (15.9%) | 8 (14.8%) | 4 (12.9%) | 6 (16.2%) | 6 (21.4%) | 5 (21.7%) |
| Age at formula introduction among combination fed subjects, wk (SD) | 20.9 (17.3) | 18.7 (16.8) | 14.7 (14.2) | 16.4 (13.7) | 17.7 (17.0) | 14.1 (15.7) |
| Gut microbiome sample age wk, mean (SD) | 6.7 (2.0) | 6.5 (1.1) | 8.0 (4.8) | 6.5 (1.0) | 7.6 (4.4) | 6.1 (0.9) |
The following variables had missing data that is not shown in the table: birthweight (n = 10), birthweight z score (n = 14), gestational diabetes (n = 21), Pre-eclampsia (n = 21), gestational hypertension (n = 20), education (n = 14), Mediterranean diet score (n = 17), smoking (n = 12), prenatal antibiotics (n = 24), age first formula (n = 113).
Overall Microbial Community Composition
Using Bray Curtis and unweighted UniFrac distances, we found that the overall microbial community composition of the 6-week stools was significantly different by pre-pregnancy BMI category after adjusting for confounders in the vaginally delivered strata (PERMANOVA p for Bray Curtis = 0.05; PERMANOVA p for unweighted Unifrac = 0.013). However, there were no differences in Bray Curtis or unweighted Unifrac by pre-pregnancy BMI in Cesarean-born infants (Bray Curtis p = 0.813; unweighted Unifrac p = 0.159). Unweighted inter and intragroup distances are displayed in Figure S2. Different associations were observed using weighted UniFrac distances (vaginal delivery p = 0.215, Cesarean delivery p = 0.003). Unlike pre-pregnancy BMI, gestational weight gain was not associated with microbial community beta diversity in vaginal or Cesarean delivered infants.
Gut Microbiota Alpha Diversity
Alpha diversity indices are used to evaluate species richness (the number of unique OTUs) within a microbiome, and how evenly distributed they are within the ecosystem (evenness is considered by the Shannon diversity and Chao1 indices). Among vaginally-delivered infants, those born to obese mothers had significantly greater alpha diversity (higher observed OTUs, p = 0.012) when compared to their normal weight counterparts, even after multivariable adjustment (Figure 1). Similar trends were found when alpha diversity was measured by the Shannon diversity index (p = 0.069) and Chao1 (p = 0.010) (Figure S3). There were no significant findings between pre-pregnancy BMI and infant gut microbiota alpha diversity in infants delivered by Cesarean section (Figure 1; Figure S3). Moreover, gestational weight gain was not found to be independently associated with microbial alpha diversity in either vaginally-delivered or Cesarean-delivered infants (Figure S4).
Figure 1. Adjusted infant gut microbial alpha-diversity by pre-pregnancy BMI category, measured by Observed OTUs.
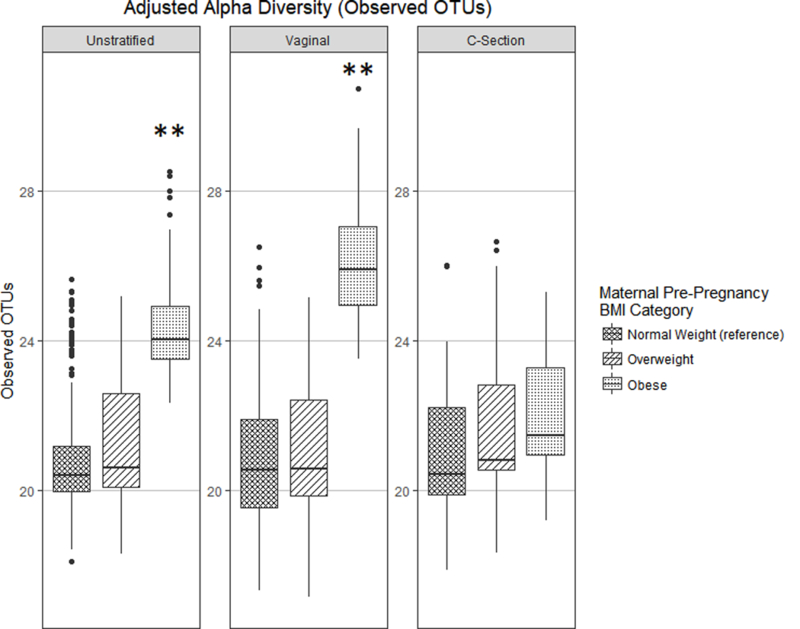
** p-value < 0.05.
Differential Abundance of Gut Microbiota
Figure 2 shows the adjusted relative abundances by pre-pregnancy BMI across delivery mode strata, with infants born to normal weight mothers as the referent group. There was a borderline significant interaction between delivery mode and pre-pregnancy BMI on our a priori hypothesized genus of interest, Bacteroides (p for interaction = 0.05). There were opposing, non-significant trends with pre-pregnancy BMI in each delivery-mode stratum (Figure 2). In vaginally-delivered infants, maternal obesity was associated with greater abundance of Parabacteroides (p = 0.03) and Staphylococcus (p = 0.022), each following a graded linear relationship with pre-pregnancy BMI categories. Furthermore, among the vaginally-delivered group, we found higher abundance of Escherichia (p < 0.001), Enterococcus (p = 0.017), Klebsiella (p = 0.037), and Ruminococcus (p < 0.001) in infants born to overweight mothers, and decreased Dorea (p = 0.043) in infants born to obese mothers. Among Cesarean-delivered infants, obesity was associated with higher levels of Staphylococcus (p = 0.022), and lower levels of Escherichia (p = 0.008).
Figure 2. Adjusted relative abundance of most abundant genera (> 1%) by pre-pregnancy BMI Category.
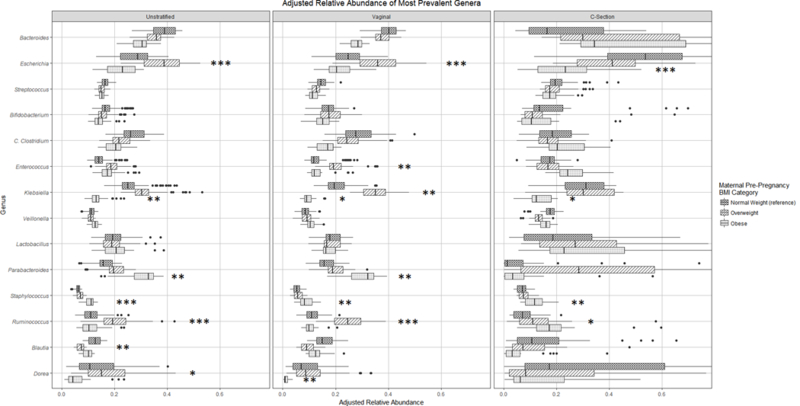
* p-value < 0.1, ** p-value < 0.05, *** p-value < 0.01.
Gestational weight gain results are shown in Figure S5, with adequate weight gain as the referent group. Among vaginally-born infants, excessive weight gain was associated with higher abundances of Escherichia (p = 0.033) and Dorea (p < 0.001). Within Cesarean-born infants, excessive weight gain was associated with lower levels of Dorea (p = 0.027) and inadequate weight gain was related to lower levels of Bifidobacterium (p = 0.027).
To agnostically test for associations with taxa lower than the genus-level, we analyzed the differential abundance of bacterial OTUs, again according to pre-pregnancy BMI and gestational weight gain categories. Infants born vaginally to overweight mothers, compared to normal weight mothers, had significantly higher abundance of OTUs in the genera Staphylcoccus and Enterococcus, as well as species V. dispar, B. fragilis, and E. coli (FDR adjusted p < 0.1) (Figure 3). There were no significant results found for pre-pregnancy BMI in the Cesarean delivered arm, nor by gestational weight gain categories in vaginally or cesarean born infants.
Figure 3. Significant OTUs that were significantly differentially abundant (FDR-adjusted p-value < 0.1).
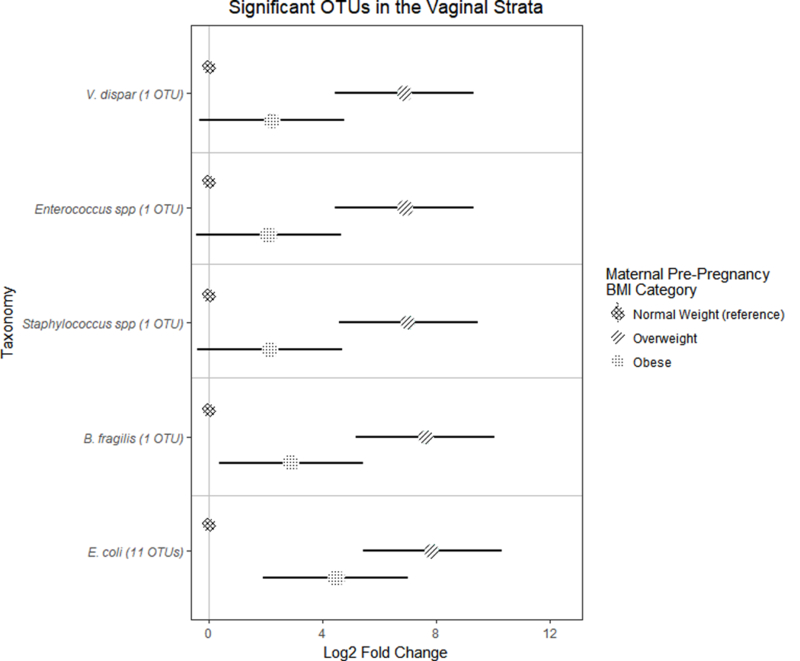
Only vaginally born infants with overweight mothers had significant differences in the abundance of OTUs, compared to normal weight mothers. Error bars indicate standard error.
Sensitivity Analyses
Our findings were not materially altered in sensitivity analyses in which we adjusted for maternal antibiotic use or feeding method at 6 weeks, nor in those in which we adjusted for sex. They were also similar when we examined associations in exclusively breastfed infants and conducted analyses separately in males and females.
Discussion
The association between pre-pregnancy BMI and several dominant taxa including Bacteroides—the most abundant genus in the infant microbiome—varied by delivery mode, often with trends in opposing directions. In vaginally born infants, pre-pregnancy BMI was associated with differences in measures of overall infant gut microbial community composition (Bray-Curtis and Unweighted Unifrac but not Weighted Unifrac) and diversity (richness and evenness), with infants born vaginally to obese mothers specifically having higher diversity. Among infants delivered vaginally, those born to overweight mothers also had higher abundance of 15 OTUs compared to those born to normal weight mothers. Pre-pregnancy BMI was not associated with microbial community diversity or differential taxa abundance in Cesarean-delivered infants. As such, our study provides further evidence that the association between maternal pre-pregnancy BMI and the infant microbiome is modified by delivery mode. We hypothesize the difference in associations by delivery mode may derive from whether an infant is exposed to maternal microbiota at birth.
Although previous studies have been conducted on the associations of pre-pregnancy BMI and gestational weight gain with the infant gut microbiome, results thus far have been conflicting. The first study on this topic was conducted in 2010 by Collado et al. in a small Finnish cohort of 42 mothers (35). In this study, infants born to overweight mothers had lower levels of Bacteroides at 1 month, and higher Staphylococcus at 6 months. Furthermore, they reported lower Bacteroides in the excess weight gain group when infants were 1 month, along with higher Staphylococcus aureus when they were 6 months. In our study, we found a similar association between pre-pregnancy BMI and Staphylococcus, but the direction of the association of pre-pregnancy BMI with infant Bacteroides (genus) varied by delivery mode and was not significant. In another study, Galley and colleagues, in 77 mothers and their infants (18—27 months) from the US, reported that maternal obesity was associated with overall infant gut microbial community composition (beta diversity), greater diversity, and higher relative abundance of Parabacteroides, findings similar to our study. However, these associations were restricted to the high socioeconomic strata (36). A more recent study of 169 maternal-infant pairs from Norway reported no associations between pre-pregnancy BMI and the overall infant gut microbial community composition in the first two years of life. Yet, similar to our study, they reported odds of having a higher prevalence of Parabacteroides in infants born to overweight or obese mothers (37).
Only two prior studies to our knowledge have assessed associations stratified by delivery mode. In 74 Brazilian mother-infant pairs (15), Mueller et al. found that pre-pregnancy BMI was associated with microbial community composition, but not alpha diversity, in vaginally-delivered infants, but not Cesarean-delivered infants. The birth-mode specific associations with overall microbial community composition were primarily driven by higher abundance of the genus Bacteroides in the infant born vaginally to overweight or obese mothers. A more recent study by Tun et al. studied associations with the infant gut microbiome at 3–4 months in a large Canadian cohort of 935 mother-infant dyads (14). In this study, the authors found that, similar to our study, maternal overweight or obesity was associated with greater infant microbial alpha diversity. Furthermore, linear discriminant analyses revealed that infants born vaginally to overweight/obese mothers had increased levels of the genus Bacteroides. The authors also reported positive associations with the Family Lachnospiraceae, which we did not observe in our study (see Supplemental Table S2). Furthermore, we were unable to replicate the relationships they observed (in their eTable3) for maternal pre-pregnancy BMI, stratified by delivery mode, with various gut microbial phyla and families (Supplemental Table S2).
While our findings are largely consistent with those from Tun et al., the instances where they diverge may derive from a variety of sources. First, unlike Tun et al., we separated overweight and obese mothers, allowing us to examine graded relationships in our analyses. Second, not only did our analytic approach (negative binomial and zero-inflated beta regression) vary from theirs (nonparametric Kruskal-Wallis test, Dunn test and linear discriminate analyses), we also controlled for confounders not accounted for in their analysis (i.e. maternal diet and maternal education). Third, population and timing of stool collection also differed between our study (assessed at infant age 6 weeks) and theirs (at age 3 – 4 months), which matters because the infant microbiome changes dynamically over the first two years of life (38). Finally, differences in DNA extraction, sequencing protocols, and pre-processing steps (we required 99% similarity to reference genomes, whereas they used 97% similarity) may contribute, in part, to the incongruent findings between our study and theirs.
As noted, our finding of higher bacterial diversity among infants born vaginally to obese mothers is consistent with two previous studies (14, 36). Galley et al. reported higher alpha diversity in infants born to overweight/obese mothers compared to normal weight mothers, but these findings were restricted to their high socioeconomic status strata (36). Tun and colleagues also found that maternal OWOB was associated with higher alpha diversity in both vaginally-delivered and Cesarean-delivered infants at 3 – 4 months of age (14). We only observed a significant association between pre-pregnancy BMI and infant microbiome diversity in the vaginally-delivered arm, although our Cesarean-born infants followed a similar but less marked, non-significant trend.
Little is known about the drivers of infant gut microbiome alpha diversity in the first months of life, nor the association of early gut microbiome diversity and childhood health status. Azad and colleagues found that formula feeding compared to breastfeeding was associated with higher alpha diversity at 3 months of life, yet lower alpha diversity (in the same infants) at 1 year (39). However, our diversity findings remained unchanged when we restricted to exclusively breastfed infants and when we adjusted for breastfeeding. We hypothesize that unlike in adulthood, where higher diversity is associated with better host health, higher microbial diversity in early infancy may indicate less mature immune system development. Large longitudinal birth cohort studies are needed to robustly examine the determinants of infant diversity in early life (first 3 months), its bi-directional relationship with immunological development, and whether it is associated with disease risk later in life.
Our finding of significantly higher B. fragilis in infants born vaginally to overweight mothers is worth highlighting, as this species may be vertically transferred from mother to newborn at birth (11) and is implicated in childhood overweight and obesity. Using culture-based techniques, a Belgian study of 138 infants found B. fragilis in infants (at 1 and 3 months age) is associated with higher BMI in later childhood (40). Furthermore, a study of 84 Brazilian children 3 – 11 years old, along with a Netherlands cohort of 909 one-month old infants with BMI-z-scores measured between 1 and 10 years of age, showed similar results (41, 42). Further studies on this species and its function are warranted to understand its potential relationship with obesity and other metabolic diseases later in life.
We also reported graded linear associations of pre-pregnancy BMI with Parabacteroides, within vaginally delivered neonates, and with the genus Staphylococcus across both strata of delivery mode. OTUs within both of these genera were also significantly higher among infants born vaginally to overweight mothers. Considering Parabacteroides and Staphylococcus have found to be related to negative health outcomes in both children (43–46) and adults (47–51), future research is needed to assess if the associations between maternal weight status and these bacterial taxa mediate the risk for dysfunctional health, including obesity, later in life.
In general, the gut microbiome of infants born vaginally to overweight and obese mothers showed a higher abundance of OTUs that are generally associated with gut dysbiosis (e.g. B. fragilis, E. coli, Enterococcus, and Staphylococcus). Although there were several genera associated with gestational weight gain, on the whole associations of gestational weight gain with infant gut microbiome signatures were less marked, and gestational weight gain was not significantly associated with measures of diversity alpha diversity in vaginally-delivered or Cesarean-delivered infants. This finding suggests that pre-pregnancy BMI is more influential than gestational weight gain on the development of the infant microbiome.
Our study has several strengths that distinguish it from previous studies in this field. The large sample size within the New Hampshire Birth Cohort Study allowed us to stratify by delivery mode, and to separate obese mothers from overweight mothers and inadequate weight gain from adequate weight gain. Comprehensive data on covariates also enabled us to adjust for confounders of interest, and to test for effect measure modification in our models. However, there are also limitations to our study. The NHBC comprises a cohort of primarily Caucasian mother-infant pairs, which may not be representative of the general U.S. population. Furthermore, because our analyses were restricted to stool samples at 6 weeks of age, our findings may not generalize to later developmental stages of infancy and childhood. Finally, our study is observational and thus we cannot rule out the possibility that unmeasured or residual confounding influenced our findings.
In conclusion, we found that maternal pre-pregnancy BMI was associated with infant microbiome diversity and composition at 6 weeks of age. Yet, importantly, these associations were largely seen only in vaginally-delivered infants, consistent with the hypothesis that delivery mode may modify the mother-to-newborn transfer of maternal obese bacteria. As the initial transfer of microbiota from mothers to their infants is critical for providing the infant with bacteria that educate the immune system and break down otherwise indigestible nutrients to make energy, vitamins and minerals, there is clear need for continued study of potential perturbations to healthy mother-to-newborn transfer of microbiota and how these impacts may aid in understanding the early origins of disease. Furthermore, studies are needed to determine how the associations with the microbiome observed in our study relate to differential risk for obesity and other health outcomes later in life.
Supplementary Material
Acknowledgements
We would like to extend our sincerest gratitude to the participants of the New Hampshire Birth Cohort. Furthermore, we would like to thank Michael S. Zens, and John Hudson for their unending patience and extensive technological help.
Funding: Research reported in this publication was supported by The Mid-Atlantic Nutrition Obesity Research Center (NORC) under NIH award number P30DK072488. Research reported in this publication was also supported by the National Heart, Lung, And Blood Institute of the National Institutes of Health under Award Number K01HL141589 (PI: Mueller), National Institutes of Health (grants NIGMS P20GM104416 (Karagas), NIEHS P01ES022832 (Karagas), NLM K01LM011985 (Hoen), NLM R01LM012723 (Hoen), the US Environmental Protection Agency (RD83544201) (Karagas). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Compliance with ethical standards
Competing Interests: The authors declare that they have no competing interests.
References
- 1.Skinner AC, Ravanbakht SN, Skelton JA, Perrin EM, Armstrong SC. Prevalence of Obesity and Severe Obesity in US Children, 1999–2016. Pediatrics. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hales CM, Fryar CD, Carroll MD, Freedman DS, Ogden CL. Trends in Obesity and Severe Obesity Prevalence in US Youth and Adults by Sex and Age, 2007–2008 to 2015–2016. JAMA. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hruby A, Hu FB. The Epidemiology of Obesity: A Big Picture. Pharmacoeconomics. 2015;33(7):673–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joint child malnutrition estimates – levels and trends. United Nations Children’s Fund, World Health Organization, The World Bank Group; 2017. [Google Scholar]
- 5.Collaboration NCDRF. Worldwide trends in body-mass index, underweight, overweight, and obesity from 1975 to 2016: a pooled analysis of 2416 population-based measurement studies in 128.9 million children, adolescents, and adults. Lancet. 2017;390(10113):2627–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naess M, Holmen TL, Langaas M, Bjorngaard JH, Kvaloy K. Intergenerational Transmission of Overweight and Obesity from Parents to Their Adolescent Offspring - The HUNT Study. PLoS One. 2016;11(11):e0166585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitaker KL, Jarvis MJ, Beeken RJ, Boniface D, Wardle J. Comparing maternal and paternal intergenerational transmission of obesity risk in a large population-based sample. Am J Clin Nutr. 2010;91(6):1560–7. [DOI] [PubMed] [Google Scholar]
- 8.Whitaker RC, Wright JA, Pepe MS, Seidel KD, Dietz WH. Predicting obesity in young adulthood from childhood and parental obesity. N Engl J Med. 1997;337(13):869–73. [DOI] [PubMed] [Google Scholar]
- 9.Llewellyn CH, Trzaskowski M, Plomin R, Wardle J. Finding the missing heritability in pediatric obesity: the contribution of genome-wide complex trait analysis. Int J Obes (Lond). 2013;37(11):1506–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wexler HM. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev. 2007;20(4):593–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bjerke GA, Wilson R, Storro O, Oyen T, Johnsen R, Rudi K. Mother-to-child transmission of and multiple-strain colonization by Bacteroides fragilis in a cohort of mothers and their children. Appl Environ Microbiol. 2011;77(23):8318–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korpela K, Costea P, Coelho LP, Kandels-Lewis S, Willemsen G, Boomsma DI, et al. Selective maternal seeding and environment shape the human gut microbiome. Genome Res. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh S, Karagas MR, Mueller NT. Charting the Maternal and Infant Microbiome: What Is the Role of Diabetes and Obesity in Pregnancy? Curr Diab Rep. 2017;17(2):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tun HM, Bridgman SL, Chari R, Field CJ, Guttman DS, Becker AB, et al. Roles of Birth Mode and Infant Gut Microbiota in Intergenerational Transmission of Overweight and Obesity From Mother to Offspring. JAMA Pediatr. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mueller NT, Shin H, Pizoni A, Werlang IC, Matte U, Goldani MZ, et al. Birth mode-dependent association between pre-pregnancy maternal weight status and the neonatal intestinal microbiome. Sci Rep. 2016;6:23133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madan JC, Hoen AG, Lundgren SN, Farzan SF, Cottingham KL, Morrison HG, et al. Association of Cesarean Delivery and Formula Supplementation With the Intestinal Microbiome of 6-Week-Old Infants. JAMA Pediatr. 2016;170(3):212–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilmore LA, Redman LM. Weight gain in pregnancy and application of the 2009 IOM guidelines: toward a uniform approach. Obesity (Silver Spring, Md). 2015;23(3):507–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.In: Rasmussen KM, Yaktine AL, editors. Weight Gain During Pregnancy: Reexamining the Guidelines. The National Academies Collection: Reports funded by National Institutes of Health; Washington (DC)2009. [PubMed] [Google Scholar]
- 19.Wu GD, Lewis JD, Hoffmann C, Chen YY, Knight R, Bittinger K, et al. Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol. 2010;10:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Degnan PH, Ochman H. Illumina-based analysis of microbial community diversity. ISME J. 2012;6(1):183–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6(8):1621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDonald D, Clemente JC, Kuczynski J, Rideout JR, Stombaugh J, Wendel D, et al. The Biological Observation Matrix (BIOM) format or: how I learned to stop worrying and love the ome-ome. Gigascience. 2012;1(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4):e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6(3):610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73(5):1576–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vazquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience. 2013;2(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jari Oksanen FGB, Michael Friendly, Roeland Kindt, Pierre Legendre, Dan McGlinn, Peter R. Minchin, R. B. O’Hara, Gavin L. Simpson, Peter Solymos, M. Henry H. Stevens, Eduard Szoecsand Helene Wagner. vegan: Community Ecology Package. R package version 24–5. 2017. [Google Scholar]
- 32.Rigby RAS DM Generalized additive models for location, scale and shape,(with discussion). Appl Statist. 2005;54(part 3):507–54. [Google Scholar]
- 33.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol. 2014;10(4):e1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collado MC, Isolauri E, Laitinen K, Salminen S. Effect of mother’s weight on infant’s microbiota acquisition, composition, and activity during early infancy: a prospective follow-up study initiated in early pregnancy. Am J Clin Nutr. 2010;92(5):1023–30. [DOI] [PubMed] [Google Scholar]
- 36.Galley JD, Bailey M, Kamp Dush C, Schoppe-Sullivan S, Christian LM. Maternal obesity is associated with alterations in the gut microbiome in toddlers. PLoS One. 2014;9(11):e113026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stanislawski MA, Dabelea D, Wagner BD, Sontag MK, Lozupone CA, Eggesbo M. Pre-pregnancy weight, gestational weight gain, and the gut microbiota of mothers and their infants. Microbiome. 2017;5(1):113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8(343):343ra82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Azad MB, Konya T, Persaud RR, Guttman DS, Chari RS, Field CJ, et al. Impact of maternal intrapartum antibiotics, method of birth and breastfeeding on gut microbiota during the first year of life: a prospective cohort study. BJOG. 2016;123(6):983–93. [DOI] [PubMed] [Google Scholar]
- 40.Vael C, Verhulst SL, Nelen V, Goossens H, Desager KN. Intestinal microflora and body mass index during the first three years of life: an observational study. Gut Pathog. 2011;3(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ignacio A, Fernandes MR, Rodrigues VA, Groppo FC, Cardoso AL, Avila-Campos MJ, et al. Correlation between body mass index and faecal microbiota from children. Clin Microbiol Infect. 2016;22(3):258 e1–8. [DOI] [PubMed] [Google Scholar]
- 42.Scheepers LE, Penders J, Mbakwa CA, Thijs C, Mommers M, Arts IC. The intestinal microbiota composition and weight development in children: the KOALA Birth Cohort Study. Int J Obes (Lond). 2015;39(1):16–25. [DOI] [PubMed] [Google Scholar]
- 43.Rogers MB, Firek B, Shi M, Yeh A, Brower-Sinning R, Aveson V, et al. Disruption of the microbiota across multiple body sites in critically ill children. Microbiome. 2016;4(1):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Collado MC, Calabuig M, Sanz Y. Differences between the fecal microbiota of coeliac infants and healthy controls. Curr Issues Intest Microbiol. 2007;8(1):9–14. [PubMed] [Google Scholar]
- 45.de Meij TG, de Groot EF, Eck A, Budding AE, Kneepkens CM, Benninga MA, et al. Characterization of Microbiota in Children with Chronic Functional Constipation. PLoS One. 2016;11(10):e0164731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wopereis H, Sim K, Shaw A, Warner JO, Knol J, Kroll JS. Intestinal microbiota in infants at high risk for allergy: Effects of prebiotics and role in eczema development. J Allergy Clin Immunol. 2018;141(4):1334–42 e5. [DOI] [PubMed] [Google Scholar]
- 47.Wang F, Yu T, Huang G, Cai D, Liang X, Su H, et al. Gut Microbiota Community and Its Assembly Associated with Age and Diet in Chinese Centenarians. J Microbiol Biotechnol. 2015;25(8):1195–204. [DOI] [PubMed] [Google Scholar]
- 48.Yan Q, Gu Y, Li X, Yang W, Jia L, Chen C, et al. Alterations of the Gut Microbiome in Hypertension. Front Cell Infect Microbiol. 2017;7:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Million M, Tidjani Alou M, Khelaifia S, Bachar D, Lagier JC, Dione N, et al. Increased Gut Redox and Depletion of Anaerobic and Methanogenic Prokaryotes in Severe Acute Malnutrition. Sci Rep. 2016;6:26051. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Drell T, Lutsar I, Stsepetova J, Parm U, Metsvaht T, Ilmoja ML, et al. The development of gut microbiota in critically ill extremely low birth weight infants assessed with 16S rRNA gene based sequencing. Gut Microbes. 2014;5(3):304–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stryjewski ME, Corey GR. Methicillin-resistant Staphylococcus aureus: an evolving pathogen. Clin Infect Dis. 2014;58 Suppl 1:S10–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


