Figure 5. Lack of Sen1 produces extended read‐through tRNA transcripts.
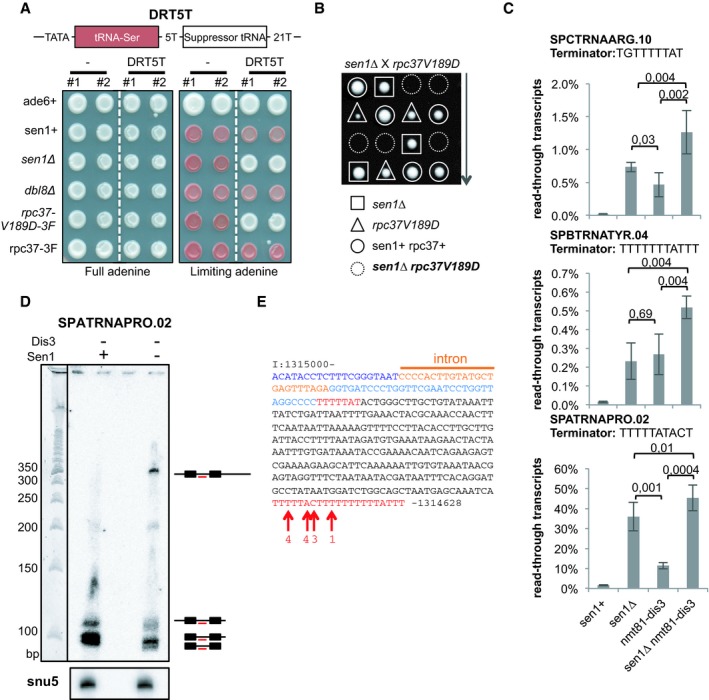
- Cells of the indicated genotypes that carried or not the DRT5T dimeric tRNA construct (schematized on top) were grown either in the presence of the optimum concentration of adenine (left) or in the presence of a limiting concentration of adenine (right). Two independent clones of the same genotype (#1 and #2) were used. See text for details. 3F refers to the 3Flag epitope tag at the C‐terminus of Rpc37.
- Tetrad dissection was used to show that the double‐mutant sen1∆ rpc37‐V189D is dead.
- Strand‐specific RT–qPCR was used to quantify the levels of read‐through transcripts (see Materials and Methods). The mean ± SD from four biological replicates is represented here. P‐values were obtained using the Wilcoxon–Mann–Whitney statistical test.
- Northern blot analysis of the tRNA SPATRNAPRO.02 using an intron‐specific probe (TCTAAACTCAGCATACAAGTGGGG). U5 snRNA was used as a loading control.
- Sequence of the ˜350‐nt‐long read‐through transcript at SPATRNAPRO.02. Residues in blue represent the sequence of the mature tRNA. Residues in red represent potential terminator sequences. Red arrows show the 3′ end nucleotide of the read‐through transcripts. The numbers indicate the number of times the sequenced transcripts terminated at the indicated position.
