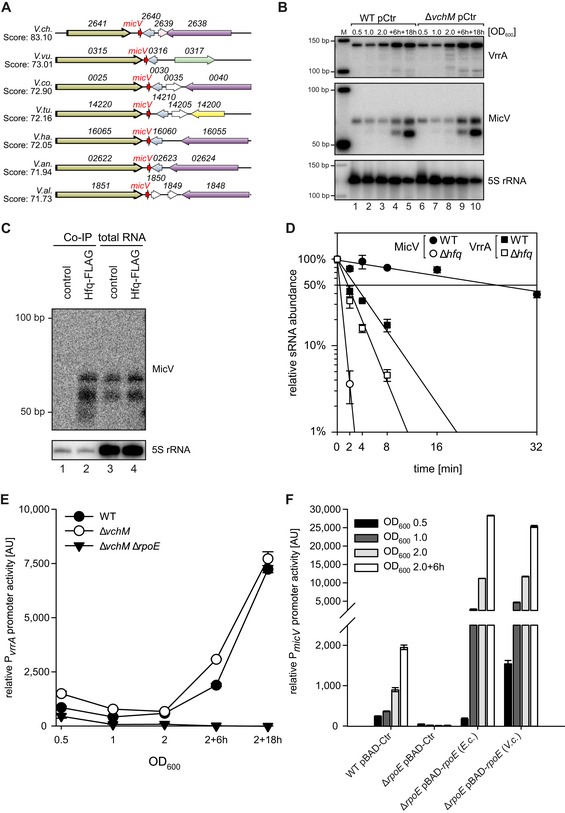
Gene synteny analysis between the genomic loci encoding micV in various Vibrio strains. Homologous genes are indicated by the same colors.
Vibrio cholerae wild‐type and ΔvchM strains carrying empty vector control plasmids (pCtr) were grown in LB medium. At the indicated time points, RNA samples were collected and tested for micV and vrrA expression by Northern blot analysis. A size marker is provided on the left (M), and 5S rRNA was used as loading control.
Vibrio cholerae wild‐type (control) and hfq::hfq‐3xFLAG (Hfq‐FLAG) strains were grown to stationary phase (OD600 of 2), lysed, and subjected to immunoprecipitation using the anti‐FLAG antibody. RNA samples of lysate (total RNA) and co‐immunoprecipitated fractions were analyzed on Northern blots. 5S rRNA served as loading control.
Vibrio cholerae wild‐type and Δhfq strains were cultivated in LB medium to an OD600 of 1.0. Cells were treated with rifampicin to terminate transcription. Total RNA samples were collected at the indicated time points, and MicV or VrrA transcript levels were monitored on Northern blots.
Vibrio cholerae wild‐type, ΔvchM, and ΔvchM ΔrpoE strains harboring PvrrA::mKate2 plasmids were grown in M9 minimal medium. Samples were collected at various stages of growth and analyzed for fluorescence.
Escherichia coli BW25113 wild‐type and ΔrpoE strains carrying PmicV::gfp plasmids and either empty vector control (pBAD‐Ctr) or plasmids expressing rpoE of E. coli (pBAD‐rpoE (E.c)) or of V. cholerae (pBAD‐rpoE (V.c)) were grown in LB medium, supplemented with L‐arabinose (0.2% final conc.). Samples were collected at various stages of growth and analyzed for fluorescence.
= 3.