Abstract
Metabolomic research has emerged as a promising approach to identify potential biomarkers in multiple sclerosis (MS). The aim of the present study was to determine the effect of interferon beta (IFN ß) on the metabolome of MS patients to explore possible biomarkers of disease activity and therapeutic response. Twenty-one MS patients starting IFN ß therapy (Rebif® 44 μg; s.c. 3 times per week) were enrolled. Blood samples were obtained at baseline and after 6, 12, and 24 months of IFN ß treatment and were analyzed by high-resolution nuclear magnetic resonance spectroscopy. Changes in metabolites were analyzed. After IFN ß exposure, patients were divided into responders and nonresponders according to the “no evidence of disease activity” (NEDA-3) definition (absence of relapses, disability progression, and magnetic resonance imaging activity), and samples obtained at baseline were analyzed to evaluate the presence of metabolic differences predictive of IFN ß response. The results of the investigation demonstrated differential distribution of baseline samples compared to those obtained during IFN ß exposure, particularly after 24 months of treatment (R2X = 0.812, R2Y = 0.797, Q2 = 0.613, p = 0.003). In addition, differences in the baseline metabolome between responder and nonresponder patients with respect to lactate, acetone, 3-OH-butyrate, tryptophan, citrate, lysine, and glucose levels were found (R2X = 0.442, R2Y = 0.768, Q2 = 0.532, p = 0.01). In conclusion, a metabolomic approach appears to be a promising, noninvasive tool that could potentially contribute to predicting the efficacy of MS therapies.
Electronic supplementary material
The online version of this article (10.1007/s13311-019-00721-8) contains supplementary material, which is available to authorized users.
Key Words: Multiple sclerosis, metabolomic analysis, biomarkers, interferon beta 1a, treatment monitoring
Introduction
Multiple sclerosis (MS) is a complex immune-mediated disease of the central nervous system with extremely heterogeneous pathogenesis [1], clinical phenotypes, and disability trajectories [2, 3]. Several factors, both physiological and pathological [4–6], are considered to have a negative influence on MS course, although mechanisms underlying this association remain largely unexplored. Conversely, disease-modifying drugs (DMDs) for MS have modified the natural history of the disease via direct effects on inflammation and disease activity [7], leading to improved outcomes principally recognized over the short term. However, response to treatment is different in each patient and is potentially influenced by many factors. Thus, it is extremely difficult to predict long-term treatment effects on MS outcomes [8, 9], in particular for drugs launched in the next few years [10].
In this context, identification of predictive biomarkers of therapeutic response and exploration of the biological mechanisms involved in this response represent crucial and novel challenges in the therapeutic decision-making process [11], increasingly oriented toward personalized medicine [12]. For this reason, “omics” approaches—genomics/epigenomics, proteomics and metabolomics—have emerged as promising tools for capturing the complexity of MS and numerous possible factors influencing its evolution [13]. In particular, metabolomics with the detailed analysis of metabolites detected in a biological system reveals the signatures of dynamic multiparametric responses to different endogenous and exogenous processes, while evaluating epigenetic modifications [14, 15]. To date, metabolomic research in MS has been largely focused on identification of potential biomarkers for diagnosis and disease monitoring [16–18], whereas little is known about the metabolomic profiles induced by DMDs and related to the therapeutic response or possible adverse effects.
Interferon beta (IFN ß) is one of the first drugs used as a first-line therapy in MS. Thus, in addition to evidence from many controlled trials and postmarketing studies, long-term data regarding effectiveness and safety are available [19, 20]. However, in clinical practice, response to IFN ß is defined exclusively through MRI features and clinical relapses [21, 22], whereas more specific biomarkers of response to this treatment are lacking.
Based on these considerations, the purpose of this study was to explore the metabolite profile of a group of naïve relapsing remitting MS patients starting IFN ß treatment, in order to determine the effect of IFN ß on the metabolome to identify predictive biomarkers of disease activity and therapeutic response. A group of healthy subjects was also included to identify possible differences in the metabolite profiles of MS patients during IFN ß treatment. Finally, we evaluated whether specific metabolomic characteristics present at baseline were able to predict the subsequent therapeutic response to IFN ß. Metabolite profiles were evaluated in blood samples from all participants using analytical high-resolution nuclear magnetic resonance (1H-NMR) spectroscopy.
Methods
Participants
The study included a group of naïve relapsing remitting MS patients (diagnosed according to the McDonald 2010 criteria [23]) and a group of demographically and ethnically matched healthy controls.
For each patient, we prospectively collected a total of 4 blood samples for 1H-NMR spectroscopy, the first one prior to starting IFN ß therapy (Rebif® 44 μg; s.c. 3 times per week) (T0) and the following samples collected at 6 months (T1), 12 months (T2), and 24 months (T3).
Adherence to IFN ß treatment was evaluated by tracking injection history using the electronic self-injection device according to manufacturer instructions (Merk Serono, Bari, Italy), and only patients with high treatment adherence (≥ 90%) were included in the metabolomic analyses.
The patients’ clinical features (disease duration and level of disability evaluated using the Expanded Disability Status Scale (EDSS) [24] and MRI data (presence of Gd-enhancing lesions)) were recorded prior to IFN ß initiation, whereas the number clinical relapses, EDSS variations, and the presence of new/enlarging T2 or T1 Gd-enhancing lesions on MRI were collected at T2 and T3. The detection of new or enlarged T2 lesions or gadolinium-enhancing lesions was evaluated using MRI after IFN ß exposure (at T2 and T3) and compared to MRI images acquired at T1. This is because residual disease activity can still occur in the first few months of treatment, in relation to the kinetics of the onset of drug action as well as to the time to reach full biologic effect.
Patients were evaluated at the end of the study and divided in two groups: responders (R) and nonresponders (NR), according to the NEDA 3 definition (absence of clinical relapses, no EDSS confirmed disability progression sustained for 6 months, and no new/enlarging T2 or T1 Gd-enhancing lesions on MRI) [25]. The two groups were compared on the bases of baseline clinical features and blood samples obtained at T0 and after IFN ß exposure were analyzed to find possible metabolomic differences related to therapeutic response and disease activity.
MS patients with other chronic comorbidities, patients reporting corticosteroid exposure in the previous 30 days, and patients taking chronic medications were excluded, and data regarding intervening medical conditions were collected. Analogously, healthy controls must have had no chronic diseases or medications, nor a family history of MS. Inclusion and exclusion criteria for MS patients and healthy controls are reported in Table 1.
Table 1.
Inclusion and exclusion criteria for MS patients and healthy controls
| MS patients | Healthy controls | ||
|---|---|---|---|
| Inclusion criteria | Exclusion criteria | Inclusion criteria | Exclusion criteria |
| Adults ≥ 18 years of age | Corticosteroids exposure in the previous 30 days | Adults ≥ 18 years of age | No family history of MS |
| MS diagnosis according to McDonald 2010 criteria | Presence of other chronic comorbidities | Presence of chronic disease | |
| Relapsing remitting course | Use of other chronic medications | Use of chronic medications | |
| Scheduled IFN ß treatment | |||
The local institutional Ethics Committee approved the study, and written informed consent was obtained from each participant prior to participation.
Sample Preparation and Acquisition
Ten milliliters of blood were collected, and the plasma was stored at − 80 °C until analysis. Plasma samples were extracted as previously described [26]. Samples were thawed and centrifuged at 2500g for 10 min at 4 °C. An 800-μl aliquot was added to 2400 μl of a solution comprised of chloroform/methanol 1:1 plus 350 μl of distilled water (D2O). Samples were vortexed for 1 min and centrifuged for 30 min at 1700g at room temperature. The water phase was concentrated overnight using a speed vacuum. The water phase was resuspended in 630 μl of D2O and 70 μl trimethylsilyl propanoic acid (TSP) 5.07 mM. TSP was added to provide an internal reference for the chemical shifts (0 ppm), and 650 μl of the solution was transferred to a 5-mm NMR tube.
Samples were analyzed using a Varian spectrometer (UNITY INOVA 500 Agilent Technologies, Inc., Santa Clara, CA, USA) that operated at 499 MHz, equipped with a 5-mm triple resonance probe with z-axis pulsed field gradients and an auto-sampler with 50 locations. Spectra were collected at 300 K with a presaturation pulse sequence to suppress residual water signals. One-dimensional 1H-NMR was recorded with a spectral width of 6000.2, a frequency of 2 Hz, an acquisition time of 1.5 s, a relaxation delay of 2 ms, and a 90° pulse of 9.2 μs with 256 scans. Each free induction decay (FID) was zero-filled to 64 k points and the spectra were manually phased and baseline corrected. The chemical shifts were referred to the internal standard, TSP (at δ = 0.0 ppm), using the MestReNova software (version 8.1, Mestrelab Research S.L.).
1H-NMR Spectroscopic Data Processing and Multivariate Analysis
The spectrum obtained by 1H-NMR analysis was divided into consecutive “bins” of 0.04 ppm. The spectral area investigated was the region between 0.6 and 9 ppm. The regions between 4.68 and 5.2 ppm and between 5.32 and 5.96 ppm were excluded to remove variations in the presaturation of residual water resonance and spectral regions of noise. To minimize the effects of different concentrations of plasma samples, the integrated area within each bin was normalized to a constant sum of 100. The final data set consisted of a 183 × 85 matrix. The columns represent the normalized area of each bin (variables), and the rows represent the samples (subjects). A multivariate statistical analysis was performed on the matrix generated using the SIMCA-P software (ver. 14.0, Umetrics, Sweden).
Data analyses were conducted on the matrix of variables after Pareto scaling, using principal component analysis (PCA), which is important for the exploration of sample distributions without classification. To identify potential outliers, the DmodX and Hotelling’s T2 tests were applied (SIMCA-P software, ver. 14.0, Umetrics, Sweden).
Partial least square (PLS-DA) and orthogonal partial least square discriminant analyses (OPLS-DA) were subsequently applied. PLS-DA and OPLS-DA maximize the discrimination between samples assigned to different classes. Variance and predictive ability (R2X, R2Y, Q [2]) were established to evaluate the suitability of the models. In addition, a permutation test (n = 400) was performed to validate the models. The scores from each OPLS-DA model were subjected to a CV-ANOVA to test for significance (p < 0.05).
The most significant variables were evaluated by the loading plot from each model and the VIP list. Metabolites were quantified by using the Chenomx NMR Suite 7.1 (Chenomx Inc., Canada) [27] and the concentrations of metabolites’ were used to conduct univariate analysis with the GraphPad Prism 7 software. Wilcoxon paired tests were used to evaluate differences between patients under treatment at different time points, whereas Mann–Whitney U tests were used to evaluate differences between patients and controls.
Metabolic pathways were built based on published literature and MetaboAnalyst 4.0 (www.metaboanalyst.ca), a web server designed to obtain comprehensive metabolomic data analysis, visualization, and interpretation [28]. For this aim, only metabolites having significantly different concentrations between the classes of patients (p < 0.05) were used.
Data Availability
The study protocol is available on request from the principal investigator. Raw data can be made available in a de-identified form on written request to the principal investigator under the condition that an additional ethics approval is obtained from the local institutional Ethics Committee. Data not published within the article will be shared on request from any qualified investigator.
Results
The study included 21 MS patients (7, 33.3% male) of which 16 (76.2%) were classified as R after 2 years of IFN ß treatment, whereas 5 patients were NR. Blood samples collected at all time points were available for 16 patients; thus, only these patients were used for analysis of metabolite profile variation during the IFN ß treatment.
At baseline, the mean age and disease duration were 38.5 ± 10.5 and 3.4 ± 2.1 years, respectively, whereas the mean EDSS was 2.9 ± 0.7. The study also included a control group of 16 subjects with comparable demographic characteristics (6, 37.5% male; mean age 37.2 ± 9.5 years) (Table 2).
Table 2.
Demographic characteristics of MS patients and healthy controls
| Baseline characteristics | MS patients (21) | Healthy controls (16) |
|---|---|---|
| Male gender | 7 (33.3%) | 6 (37.5%) |
| Age (mean years) | 38.5 ± 10.5 | 37.2 ± 9.5 |
| MS duration (mean years) | 3.4 ± 2.1 | na |
| EDSS score (mean) | 2.9 ± 0.7 | na |
na = not applicable
Comparing R and NR patients, no differences in baseline clinical features were found, whereas gadolinium-enhancing lesions were observed using MRI at baseline in 3/5 (60%) NR patients vs 5/16 (31%) R patients as reported in Table 3.
Table 3.
Baseline characteristics of responder (R) and nonresponder (NR) patients
| Baseline characteristics | R patients (16) | NR patients (5) |
|---|---|---|
| Male gender | 5 (31.2%) | 2 (40%) |
| Age (mean years) | 38.2 ± 10.3 | 39.5 ± 10.6 |
| MS duration (mean years) | 3.3 ± 2.1 | 3.5 ± 2.2 |
| EDSS score (mean) | 2.8 ± 0.7 | 3.0 ± 0.9 |
| MRI activity (Gd + lesions) | 5 (31%) | 3 (60%) |
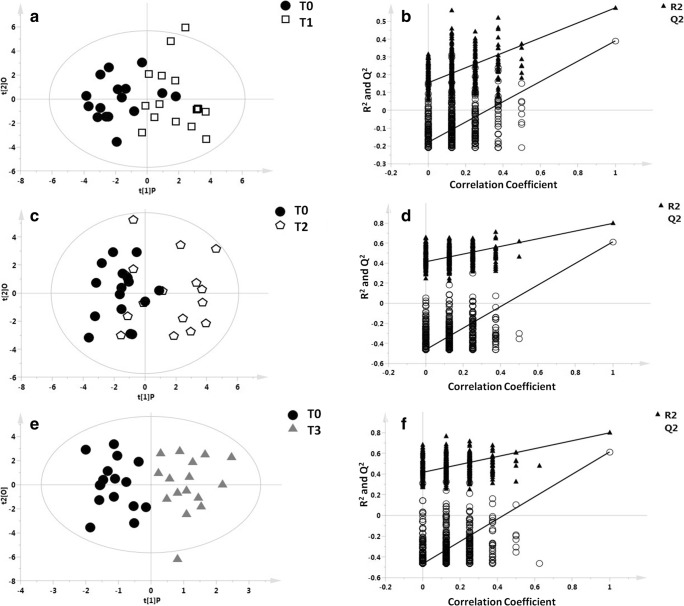
Figure 1 shows OPLS-DA models with the respective permutation tests obtained by comparing each patient’s baseline (T0) samples with their own subsequent time point samples (T0 vs T1 (months 6), T0 vs T2 (months 12), T0 vs T3 (months 24)). These models identified a differential distribution of T0 vs T1 (R2X = 0.697, R2Y = 0.576, Q2 = 0.391, p = 0.006; Fig. 1A, B) and T0 vs T3 samples (R2X = 0.812, R2Y = 0.797, Q2 = 0.613, P = 0.003; Fig. 1E, F), whereas the second model comparing each patient’s T0 vs T2 samples, although explicative, was only found to have a trend toward association (R2X = 0.725, R2Y = 0.450, Q2 = 0.247, p = 0.09; Fig. 1C, D).
Fig. 1.
OPLS-DA models with the respective permutation tests obtained by comparing T0 vs T1, T0 vs T2, and T0 vs T3 samples for each patient. Blood samples were collected at baseline (T0) and then at 6 (T1), 12 (T2), and 24 (T3) months of IFN ß treatment. OPLS-DA models with the respective permutation test indicated a differential distribution of the T0 vs T1 sample (A, B), R2X = 0.697, R2Y = 0.576, Q2 = 0.391, p = 0.006); T0 vs T2 sample (C, D), R2X = 0.725, R2Y = 0.450, Q2 = 0.247, p = 0.09); and T0 vs T3 sample (E, F), R2X = 0.812, R2Y = 0.797, Q2 = 0.613, p = 0.003)
Figure 2 shows an OPLS-DA model with 3 classes, including the T0 and T3 samples of each patient and the samples from the recruited healthy controls. This model indicates that the metabolomic profile of patients exposed to IFN ß treatment after 24 months is different from the basal metabolomic profile and more similar to the healthy control profile (R2X = 0.615, R2Y = 0.598, Q2 = 0.149, p = 0.1), although constituting a distinct class as shown by the distribution of samples in the scores plot.
Fig. 2.
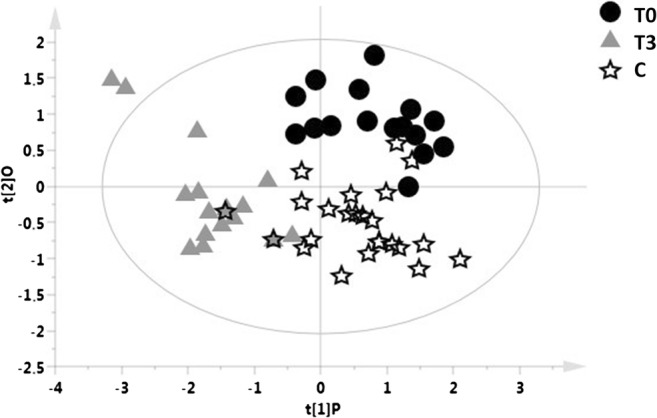
OPLS-DA model with 3 classes, including the T0 and T3 samples of each patient and the samples from healthy controls. The blood samples included in the OPLS-DA model were collected at baseline (T0) and 24 months (T3) of IFN ß treatment. OPLS-DA model of T0 (black circles) and T3 samples (gray triangles) of each patient and control group (white stars) is reported (R2X = 0.615, R2Y = 0.598, Q2 = 0.149, p = 0.1)
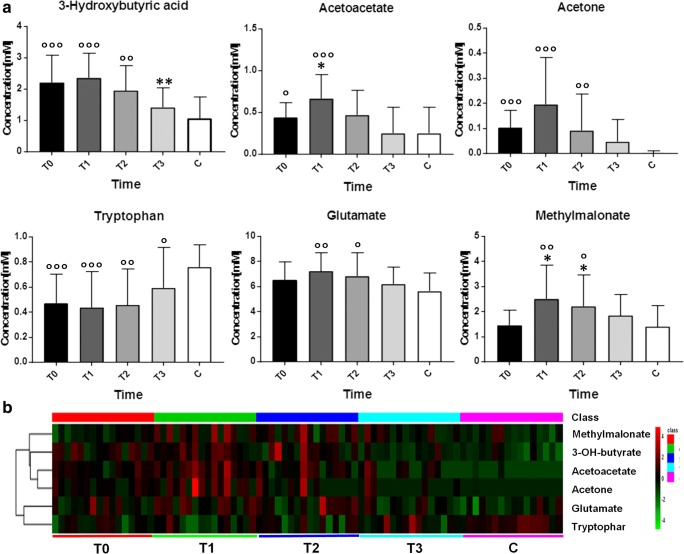
The mean concentrations of the metabolites found to be discriminants from the OPLS-DA models comparing T0 vs T1, T0 vs T2, T0 vs T3, and the control group including the variation (increase, reduction, and stability) with respect to T0 are reported in Table 4. In particular, acetoacetate, acetone, 3-hydroxybutyrate, glutamate, and methylmalonate levels significantly decreased during treatment, whereas tryptophan levels increased. The graphical representation of the variation of these putative metabolites at different time points and the heat-map of their levels during the IFN ß treatment are reported in Fig. 3, along with data from healthy controls. As shown, the variation in these metabolites was time-dependent during the IFN ß exposure, with maximum metabolic transition observed at T1 and variation in the direction of metabolite levels of healthy controls at T3.
Table 4.
Concentrations of the metabolite discriminants for the models T0 vs T1, T0 vs T2, T0 vs T3, T0 vs HC, and trend of metabolite variations compared to T0
| Metabolites | Classes | |||||||
|---|---|---|---|---|---|---|---|---|
| T1 | T2 | T3 | C | |||||
| 1,3-Dymethylurate | 0.47 | – | 0.54 | + | 0.66 | + | 0.62 | + |
| 2-Oxoglutarate | 0.99 | + | 0.86 | + | 0.64 | – | 0.77 | = |
| 3-Hydroxybutyrate | 2.36 | + | 2.23 | = | 1.35 | – | 1.66 | – |
| Acetate | 1.17 | = | 1.18 | = | 1 | = | 0.96 | = |
| Acetoacetate | 0.66 | + | 0.44 | = | 0.27 | – | 0.05 | – |
| Acetone | 0.19 | – | 0.08 | – | 0.04 | – | 0.002 | – |
| Citrate | 2.29 | = | 2.30 | = | 2.22 | = | 2.00 | = |
| Fructose | 4.70 | = | 5.18 | + | 5.96 | + | 5.29 | + |
| Glucose | 34.99 | = | 35.86 | = | 36.12 | = | 35.76 | = |
| Glutamate | 7.22 | + | 6.96 | + | 6.04 | – | 5.49 | – |
| Glycerol | 3.48 | + | 3.07 | + | 3.35 | + | 3.12 | + |
| Lactate | 25.53 | – | 26.31 | + | 26.34 | + | 28.62 | + |
| Lysine | 2.71 | + | 1.96 | – | 2.22 | = | 2.53 | = |
| Malonate | 0.73 | = | 0.65 | – | 0.86 | + | 0.36 | – |
| Scyllo-inositol | 2.96 | = | 2.60 | – | 3.05 | = | 3.02 | + |
| Methylmalonate | 2.51 | + | 2.28 | + | 1.83 | + | 1.40 | = |
| Pyroglutamate | 1.74 | + | 2.04 | + | 2.06 | + | 2.00 | + |
| Taurine | 2.06 | – | 2.38 | = | 2.45 | + | 3.07 | + |
| Tryptophan | 0.44 | = | 0.48 | = | 0.58 | + | 0.72 | + |
| Myo-inositol | 2.57 | + | 2.49 | + | 2.8 | + | 2.52 | + |
The concentrations of the different metabolites are reported in millimoles (mM). The patients’ blood samples were collected at baseline (T0) and then at 6 (T1), 12 (T2), and 24 (T3) months of IFN ß treatment. Acetoacetate, acetone, 3-hydroxybutyrate, glutamate, methylmalonate, and tryptophan were the metabolites that were changed following treatment, with p value < 0.05. All metabolites underwent univariate analysis with the Wilcoxon test, whereas the mean of each metabolite at the different time points was compared to the control group using Mann–Whitney U tests
Fig. 3.
The most important metabolites identified by the analysis of the multivariate models (T0 vs T1, T0 vs T2, and T0 vs T3) and the control group. (A) The variation of discriminant metabolites (increase, reduction, and stability) in all classes under investigation. Stars represent significant variations of concentrations compared to T0 (calculated by Wilcoxon test, a paired nonparametric statistical test) whereas circles indicate significant variation compared to C group (calculated by Mann–Whitney U test). (B) Heat-map representing data in the form of a map in which the concentrations of putative metabolites are reported with different colors for MS patients and healthy controls, at different time points (red color indicates higher levels, whereas green color lower levels)
All of these metabolites were used to investigate alterations in metabolic pathways during treatment with IFN ß, revealing that energetic pathways and tryptophan metabolism were the most relevant altered pathways.
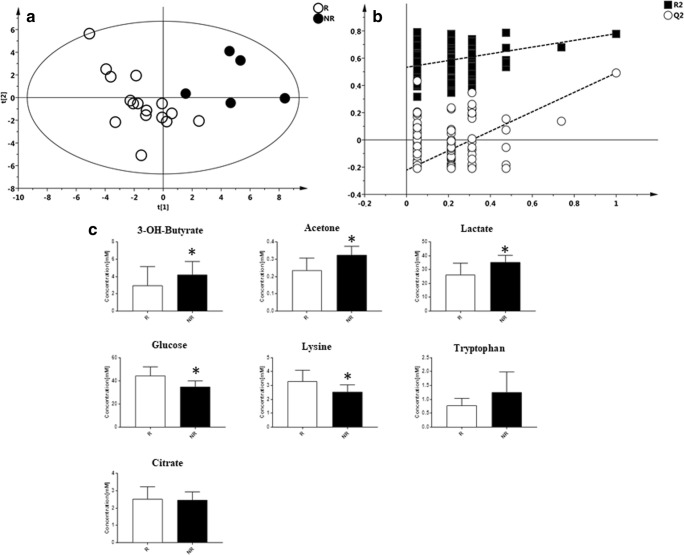
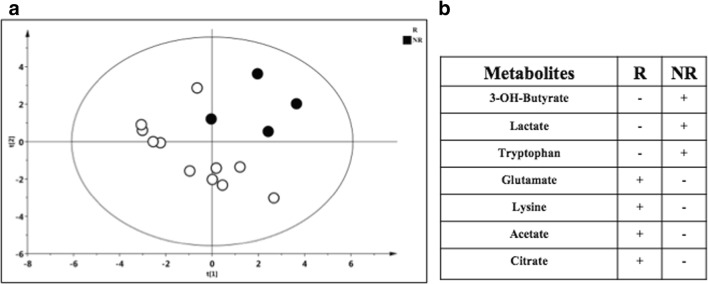
Subsequently, we investigated if the metabolic profile prior to IFN ß initiation could predict the subsequent therapeutic response. For this study, a supervised model was generated to compare T0 samples of both R and NR patients. As reported in Fig. 4A, the OPLS-DA model validated with the respective permutation test (Fig. 4B) showed a different distribution of the R vs NR patients (R2X = 0.442; R2Y = 0.768, Q2 = 0.532, p = 0.01) with 7 metabolites primarily driving the separation between the two classes (Table 5). In particular, lactate, acetone, 3-OH-butyrate, and tryptophan were higher in NR patients, whereas citrate, lysine, and glucose were higher in R patients (Fig. 4C). Finally, a supervised model was generated to compare the metabolomic profile of R and NR patients after at least 1 year of IFN ß exposure (samples available for 15 patients: 11 R and 4 NR). The model, although not significant (R2X = 0.601, R2Y = 0.696, Q2 = 0.226, p = 0.1) (Fig. 5A), indicated a separation between the two classes, with lactate, 3-OH-butyrate, and tryptophan higher in NR patients, whereas glutamate, lysine, acetate, and citrate were higher in R patients (Fig. 5B).
Fig. 4.
PLS-DA model with the respective permutation tests obtained by comparing the T0 samples of R vs NR patients to IFN ß treatment. (A) PLS-DA model obtained from 21 plasma samples of MS patients at time point T0. Patients were classified as responders (R) and nonresponders (NR) after IFN ß treatment, according to NEDA 3 definition. Statistical parameters were R2X = 0.440, R2Y = 0.768, Q2 = 0.532, p = 0.01. (B) Validation of the model by permutation test. (C) Bar graph of the most important metabolites resulting from the multivariate model of R and NR patients
Table 5.
Discriminant metabolites from the PLS-DA model of R vs NR patients at T0
| Metabolite | R | NR | ||
|---|---|---|---|---|
| Trend | Mean (mM) | Trend | Mean (mM) | |
| Lactate | – | 26.48 | + | 35.48 |
| 3-OH-Butyrate | – | 2.9 | + | 4.2 |
| Acetone | – | 0.24 | + | 0.33 |
| Tryptophan | – | 0.77 | + | 1.26 |
| Citrate | + | 2.54 | – | 2.47 |
| Lysine | + | 3.3 | – | 2.5 |
| Glucose | + | 44.37 | – | 34.97 |
Fig. 5.
OPLS-DA model obtained by comparing R vs NR patients after IFN ß exposure and metabolites driving the two classes of separation. (A) PLS-DA model obtained from 15 plasma samples of MS patients classified as responders (R) and nonresponders (NR) during IFN ß exposure (after at least 1 year), according to NEDA 3 definition. Statistical parameters were R2X = 0.601, R2Y = 0.696, Q2 = 0.226, p = 0.1, with 7 metabolites primarily driving the separation between the two classes. (B) Lactate, 3-OH-butyrate, and tryptophan were higher in NR patients, whereas citrate, lysine, acetate, and glutamate in R patients. For one NR patient, no blood samples after the first year of IFN ß exposure were available
Discussion
Previous studies using 1H-NMR spectroscopy have indicated that the plasma concentrations of some metabolites, particularly those involved in energy and tryptophan metabolism, are able to discriminate MS patients from healthy controls [17]. Similarly, some evidence has suggested that serum metabolite profiles can distinguish patients with different subtypes and stages of MS, potentially identifying those with a prevalent inflammatory course [16]. However, how metabolomic changes are associated with MS can be modified by exposure to different DMDs, and whether these metabolomic variations might reflect expression of modified disease activity it is not yet known. Based on these considerations, we performed a metabolomic study with pattern recognition methods on blood from MS patients starting IFN ß treatment (Rebif® 44 μg; s.c. 3 times per week) in order to capture the IFN ß signature on patients’ metabolite profiles and the metabolites potentially involved in the therapeutic response. In particular, comparing the baseline samples with the samples at different time points, we found that acetoacetate, acetone, 3-hydroxybutyrate, glutamate, methylmalonate, and tryptophan were metabolites that drove the OPLS-DA class separation, possibly related to the effects of IFN ß. Analyzing the metabolic trend diagrams of these metabolites, it is notable that the greatest variations occur in the first 6 months of therapy, whereas at the second year of treatment, the levels of these metabolites are similar to those of healthy controls. Interestingly, the metabolites that change during IFN ß exposure are the same metabolites that were found to discriminate the classes of MS patients and healthy controls in our previous metabolomic study [17]. Therefore, we hypothesize that interferon acts directly on the MS pathological pathways, resulting in metabolomic profiles similar to those of healthy controls. A summary of the most important pathways altered during treatment with IFN ß is shown in Fig. 6.
Fig. 6.
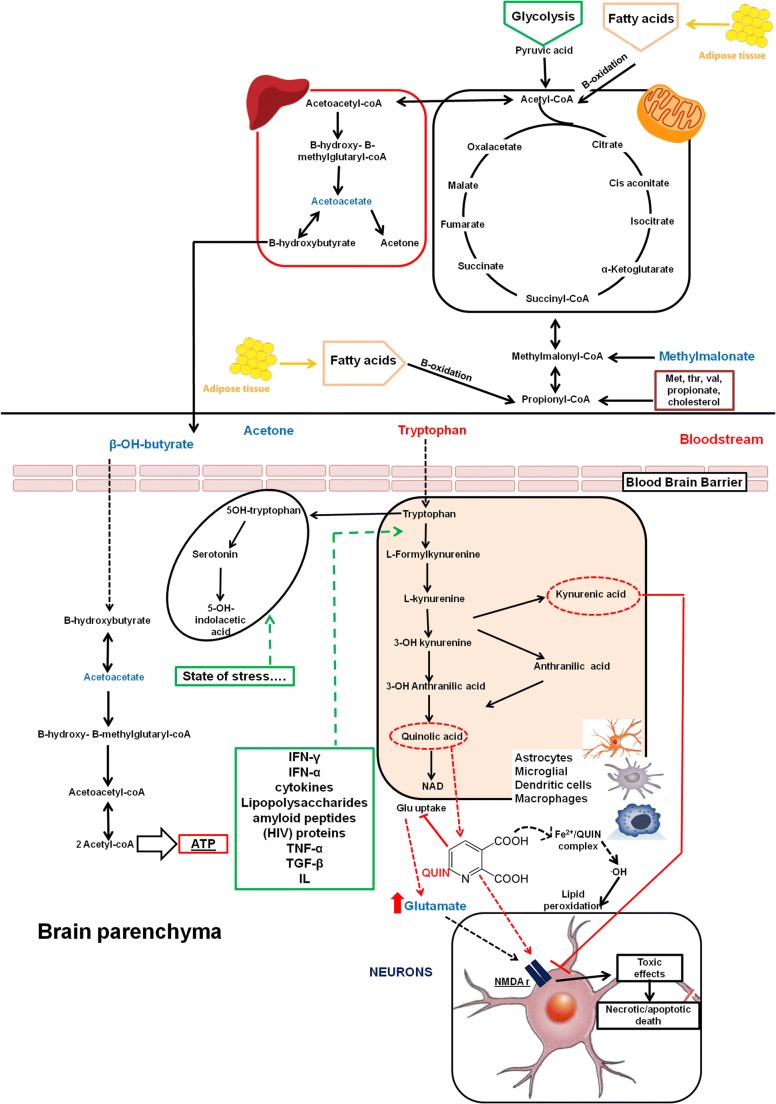
Summary of the most important pathways altered during the treatment with IFN ß. Metabolites written in blue letters were found to be increased at the beginning of treatment, whereas red metabolites were found to be decreased at the beginning of the treatment. In the case of low glucose levels, the β-oxidation of fatty acids in mitochondria is induced, resulting in production of ketone bodies (3-OH-butyrate, acetoacetate, and acetone). Ketone bodies can migrate into the bloodstream and thus are metabolized to produce ATP, especially 3-OH-butyrate, in several organs such as the brain. Tryptophan is catabolized via the kynurenine pathway in both the CNS and periphery, producing many neuroprotective (kynurenic acid and picolinic acid) and neurotoxic (3-hydroxykynurenine and quinolinic acid) metabolites
Recently, growing interest has been directed to the study of tryptophan metabolism. Several years ago, low levels of tryptophan were documented in the serum and cerebrospinal fluid of MS patients [29], suggesting its possible role in MS pathology. Principally catabolized via multistep reactions of the kynurenine pathway, tryptophan may produce many neuroactive metabolites with neuroprotective (kynurenic acid and picolinic acid) and neurotoxic (3-hydroxykynurenine and quinolinic acid) effects (as shown in Fig. 6). Tryptophan catabolism occurs in both the central nervous system and the periphery and is regulated by complex and dynamic mechanisms involving inflammatory mediators during the immune response [30]. Recently, evidence has indicated that kynurenine dysregulation is most prominent during the acute phases of MS, whereas kynurenine metabolite levels in MS patients in remission are similar to those of controls [31]. In particular, inhibition of the first enzyme that converts tryptophan to kynurenine metabolite (indoleamine 2,3-dioxygenase) seems to be associated with the reduction of disease activity [32], evidence in line with our findings documenting an increase in tryptophan level during IFN ß treatment. Nevertheless, how IFN ß dynamically influences different levels of the kynurenine pathway has not been adequately explored. Intriguingly, the levels of tryptophan in our study distinguished the R vs NR patients at baseline, highlighting the role of IFN ß treatment on this pathway. However, in the complex course of neuroinflammation in MS, the kynurenine pathway is considered a double-edged sword [33, 34]. It also contributes to neurodegeneration in the long-term due to quinolinic acid toxicity and has direct effects on glutamate uptake into astrocytes [35]. Glutamate has a plethora of effects mediated not only by a large family of receptors expressed by most cells of the nervous system, but also by many nonneural cells such as immune cells. Thus, abnormal glutamate levels in the nervous system, the direct activation of T cells by glutamate, and glutamate release by T cells can all contribute to MS-related inflammation [36]. Interestingly, at different time points of IFN ß treatment, we found a reduction in serum glutamate concentrations compared to baseline, as shown in Table 4. In addition, we found that the IFN ß exposure appears to influence levels of OH-butyrate, acetoacetate, and acetone, mainly produced in conditions of increased catabolism such as in inflammatory states, with progressive decreases in these metabolites during treatment. In addition, these energy metabolites seem to play a key role in differentiating the R vs NR patients at baseline, indicating that higher inflammatory states are a negative prognostic factor for MS course. Analogously, after IFN ß exposure, higher lactate, 3-OH-butyrate, and tryptophan levels were reported in NR patients, suggesting a possible connection of these metabolites with persistence of disease activity.
Notoriously, inflammatory conditions are related to a high demand of the organism for energy and rapid utilization of glucose, with possible increases in serum concentration of ketone bodies (e.g., butyrate and acetate). These, in addition to being used as an alternative source of energy, may also downregulate inflammatory status. In particular, butyrate has been shown to modulate proliferative T cell responses by direct interference with cell cycle progression and alteration of the stimulatory function of antigen presenting cells. In addition, butyrate has been shown to inhibit NF-kB signaling, with effects on the expression of various cytokines and cell adhesion molecules involved in immune and inflammatory responses [37, 38]. Thus, on the one hand, there was a reduction in OH-butyrate during IFN ß treatment in R patients, whereas on the other hand, the persistence of high concentrations of OH-butyrate in NR patients was associated with more marked inflammatory activity, making this metabolite an interesting biomarker of disease activity. However, the secondary down-regulatory effects of this metabolite on the immune system, as well as on the pathogenic mechanisms of MS and disease activity, still need to be explored.
Our study has several limitations to consider. First, these are preliminary data obtained in a small sample of MS patients, with the limitation that we have evaluated plasma concentrations of metabolites, whereas MS is an organ-specific disease involving exclusively the central nervous system [39]. Second, metabolomic analysis was performed exclusively using the 1H-NMR spectroscopy, and future studies with mass spectrometry may be useful to better explore the complexity of the metabolome. Finally, although subjects with other chronic medical conditions have not been recruited, we cannot exclude the possibility that the metabolites identified as discriminants are confounded by factors unrelated to MS, such as diet or other concomitant medications [40].
Conclusion
Our data suggest that treatment with IFN ß influences both energy and tryptophan metabolism, indicating that pretreatment levels of some metabolites in these pathways may influence subsequent treatment response. Considering the complexity of MS management in the current treatment era [41], 1H-NMR spectroscopy-based metabolomic analysis of blood appears to be a promising, noninvasive approach to contribute to predicting the efficacy of MS therapies, with possible implications for future personalized therapeutic decision-making processes [42]. However, our results need to be replicated in larger samples.
Electronic Supplementary Material
(PDF 14524 kb)
Acknowledgments
The authors thank Merck Serono for the financial support.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Compliance with Ethical Standards
The local institutional Ethics Committee approved the study, and written informed consent was obtained from each participant prior to participation.
Conflict of Interest
L. Lorefice received a speaker fee from Teva and serves on scientific advisory boards for Merck Serono and Biogen.
G. Fenu received honoraria for consultancy from Novartis and for speaking from Merck Serono, Biogen, and Teva.
J. Frau serves on scientific advisory boards for Biogen and Genzyme and received honoraria for speaking from Merck Serono and Teva.
G. Coghe received speaker fees from Teva and Almirall.
A. Visconti is a full time employee to Merck Serono.
L. Atzori, F. Murgia, M. Murru, and S. Tranquilli have nothing to disclose.
M. Marrosu and E. Cocco have received honoraria for consultancy or speaking from Bayer, Biogen, Novartis, Sanofi, Genzyme, Serono, and Teva.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lorena Lorefice and Federica Murgia contributed equally to this work as first authors.
Maria Giovanna Marrosu, Luigi Atzori and Eleonora Cocco equally contributed to this work as last authors.
References
- 1.Lucchinetti CF, Brück W, Rodriguez M, et al. Distinct patterns of multiple sclerosis pathology indicate heterogeneity on pathogenesis. Brain Pathol. 1996;6(3):259–74. doi: 10.1111/j.1750-3639.1996.tb00854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lublin FD, Reingold SC, Cohen JA, et al. Defining the clinical course of multiple sclerosis: the 2013 revisions. Neurology 2014 83(3):278–286 [DOI] [PMC free article] [PubMed]
- 3.Hum S, Lapierre Y, Scott SC, et al. Trajectory of MS disease course for men and women over three eras. Mult Scler. 2017;23(4):534–545. doi: 10.1177/1352458516655478. [DOI] [PubMed] [Google Scholar]
- 4.Hughes SE, Spelman T, Gray OM, et al. Predictors and dynamics of postpartum relapses in women with multiple sclerosis. Mult Scler. 2014;20(6):739–46. doi: 10.1177/1352458513507816. [DOI] [PubMed] [Google Scholar]
- 5.Zhang T, Tremlett H, Zhu F, et al. Effects of physical comorbidities on disability progression in multiple sclerosis. Neurology. 2018;90(5):e419–e427. doi: 10.1212/WNL.0000000000004885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lorefice L, Fenu G, Pitzalis R, et al. Autoimmune comorbidities in multiple sclerosis: what is the influence on brain volumes? A case-control MRI study. J Neurol. 2018;265(5):1096–1101. doi: 10.1007/s00415-018-8811-1. [DOI] [PubMed] [Google Scholar]
- 7.Confavreux C, Vukusic S. Natural history of multiple sclerosis: a unifying concept. Brain. 2006;129(Pt 3):606–16. doi: 10.1093/brain/awl007. [DOI] [PubMed] [Google Scholar]
- 8.Steyerberg EW, Claggett B. Towards personalized therapy for multiple sclerosis: limitations of observational data. Brain. 2018;141(5):e38. doi: 10.1093/brain/awy055. [DOI] [PubMed] [Google Scholar]
- 9.Sormani MP. Prognostic factors versus markers of response to treatment versus surrogate endpoints: three different concepts. Mult Scler. 2017;23(3):378–381. doi: 10.1177/1352458516676899. [DOI] [PubMed] [Google Scholar]
- 10.Cree BA, Gourraud PA, Oksenberg JR, et al. Long-term evolution of multiple sclerosis disability in the treatment era. Ann Neurol. 2016;80(4):499–510. doi: 10.1002/ana.24747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hegen H, Auer M, Deisenhammer F. Predictors of response to multiple sclerosis. Therapeutics in individual patients. Drugs. 2016;76(15):1421–1445. doi: 10.1007/s40265-016-0639-3. [DOI] [PubMed] [Google Scholar]
- 12.Jain KK. Personalized neurology. Per Med. 2005;2(1):15–21. doi: 10.1517/17410541.2.1.15. [DOI] [PubMed] [Google Scholar]
- 13.Villoslada P, Baranzini S. Data integration and systems biology approaches for biomarker discovery: challenges and opportunities for multiple sclerosis. J Neuroimmunol. 2012;248(1–2):58–65. doi: 10.1016/j.jneuroim.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 14.Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29(11):1181–9. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 15.Bhargava P, Calabresi PA. Metabolomics in multiple sclerosis. Mult Scler. 2016;22(4):451–60. doi: 10.1177/1352458515622827. [DOI] [PubMed] [Google Scholar]
- 16.Dickens AM, Larkin JR, Griffin JL, et al. A type 2 biomarker separates relapsing-remitting from secondary progressive multiple sclerosis. Neurology. 2014;83(17):1492–9. doi: 10.1212/WNL.0000000000000905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cocco E, Murgia F, Lorefice L, et al. (1)H-NMR analysis provides a metabolomic profile of patients with multiple sclerosis. Neurol Neuroimmunol Neuroinflamm. 2015;3(1):e185. doi: 10.1212/NXI.0000000000000185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poddighe S, Murgia F, Lorefice L, et al. Metabolomic analysis identifies altered metabolic pathways in multiple sclerosis. Int J Biochem Cell Biol. 2017;93:148–155. doi: 10.1016/j.biocel.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 19.Goodin DS, Reder AT, Traboulsee AL, et al. Predictive validity of NEDA in the 16- and 21-year follow-up from the pivotal trial of interferon beta-1b. Mult Scler. 2018 May 1:1352458518773511. [DOI] [PubMed]
- 20.Trojano M, Pellegrini F, Paolicelli D, et al. Real-life impact of early interferon beta therapy in relapsing multiple sclerosis. Ann Neurol. 2009;66(4):513–20. doi: 10.1002/ana.21757. [DOI] [PubMed] [Google Scholar]
- 21.Sormani MP, De Stefano N. Defining and scoring response to IFN-β in multiple sclerosis. Nat Rev Neurol. 2013;9(9):504–12. doi: 10.1038/nrneurol.2013.146. [DOI] [PubMed] [Google Scholar]
- 22.Giovannoni G, Tomic D, Bright JR, et al. “No evident disease activity”: the use of combined assessments in the management of patients with multiple sclerosis. Mult Scler. 2017;23(9):1179–118. doi: 10.1177/1352458517703193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Polman CH, Reingold SC, Banwell B, et al. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol. 2011;69(2):292–302. doi: 10.1002/ana.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurtzke JF. Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS) Neurology. 1983;33(11):1444–52. doi: 10.1212/WNL.33.11.1444. [DOI] [PubMed] [Google Scholar]
- 25.Banwell B, Giovannoni G, Hawkes C, et al. Editors’ welcome and a working definition for a multiple sclerosis cure. Mult Scler Relat Disord. 2013;2(2):65–7. doi: 10.1016/j.msard.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Murgia F, Svegliati S, Poddighe S, et al. Metabolomic profile of systemic sclerosis patients. Sci Rep. 2018;8(1):7626. doi: 10.1038/s41598-018-25992-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weljie AM, Newton J, Mercier P, et al. Targeted profiling: quantitative analysis of 1H NMR metabolomics data. Anal Chem. 2006;78(13):4430–42. doi: 10.1021/ac060209g. [DOI] [PubMed] [Google Scholar]
- 28.Xia J, Sinelnikov IV, Han B, et al. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Res. 2015;43(W1):W251–7. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monaco F, Fumero S, Mondino A, et al. Plasma and cerebrospinal fluid tryptophan in multiple sclerosis and degenerative diseases. J Neurol Neurosurg Psychiatry. 1979;42(7):640–1. doi: 10.1136/jnnp.42.7.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moffett JR, Namboodiri MA. Tryptophan and the immune response. Immunol Cell Biol. 2003;81(4):247–65. doi: 10.1046/j.1440-1711.2003.t01-1-01177.x. [DOI] [PubMed] [Google Scholar]
- 31.Aeinehband S, Brenner P, Ståhl S, et al. Cerebrospinal fluid kynurenines in multiple sclerosis; relation to disease course and neurocognitive symptoms. Brain Behav Immun. 2016;51:47–55. doi: 10.1016/j.bbi.2015.07.016. [DOI] [PubMed] [Google Scholar]
- 32.Rajda C, Majláth Z, Pukoli D, et al. Kynurenines and multiple sclerosis: the dialogue between the immune system and the central nervous system. Int J Mol Sci. 2015;16(8):18270–82. doi: 10.3390/ijms160818270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lovelace MD, Varney B, Sundaram G, et al. Current evidence for a role of the kynurenine pathway of tryptophan metabolism in multiple sclerosis. Front Immunol. 2016;7:246. doi: 10.3389/fimmu.2016.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwidzinski E, Bechmann I. IDO expression in the brain: a double-edged sword. J Mol Med (Berl). 2007;85(12):1351–9. doi: 10.1007/s00109-007-0229-7. [DOI] [PubMed] [Google Scholar]
- 35.Tavares RG, Tasca CI, Santos CE, et al. Quinolinic acid stimulates synaptosomal glutamate release and inhibits glutamate uptake into astrocytes. Neurochem Int. 2002;40(7):621–7. doi: 10.1016/S0197-0186(01)00133-4. [DOI] [PubMed] [Google Scholar]
- 36.Levite M. Glutamate, T cells and multiple sclerosis. J Neural Transm (Vienna). 2017;124(7):775–798. doi: 10.1007/s00702-016-1661-z. [DOI] [PubMed] [Google Scholar]
- 37.Böhmig GA, Krieger PM, Säemann MD, et al. n-Butyrate downregulates the stimulatory function of peripheral blood-derived antigen-presenting cells: a potential mechanism for modulating T-cell responses by short-chain fatty acids. Immunology. 1997;92(2):234–43. doi: 10.1046/j.1365-2567.1997.00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baeuerle PA, Henkel T. Function and activation of NF-kappa B in the immune system. Annu Rev Immunol. 1994;12:141–79. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 39.Steinman L. Multiple sclerosis: a two-stage disease. Nat Immunol. 2001;2(9):762–4. doi: 10.1038/ni0901-762. [DOI] [PubMed] [Google Scholar]
- 40.Sato S, Parr EB, Devlin BL, et al. Human metabolomics reveal daily variations under nutritional challenges specific to serum and skeletal muscle. Mol Metab. 2018;16:1–11. doi: 10.1016/j.molmet.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chitnis T, Giovannoni G, Trojano M. Complexity of MS management in the current treatment era. Neurology. 2018;90(17):761–762. doi: 10.1212/WNL.0000000000005399. [DOI] [PubMed] [Google Scholar]
- 42.Giovannoni G. Personalized medicine in multiple sclerosis. Neurodegener Dis Manag. 2017;7(6s):13–17. doi: 10.2217/nmt-2017-0035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 14524 kb)
Data Availability Statement
The study protocol is available on request from the principal investigator. Raw data can be made available in a de-identified form on written request to the principal investigator under the condition that an additional ethics approval is obtained from the local institutional Ethics Committee. Data not published within the article will be shared on request from any qualified investigator.