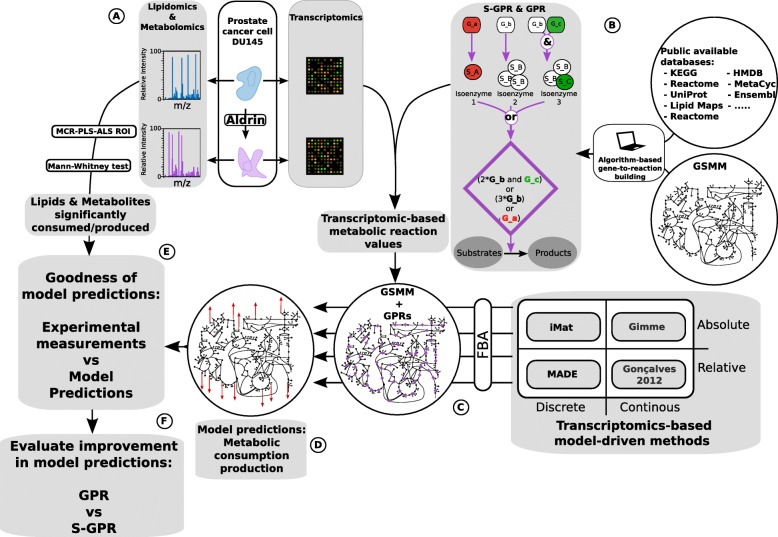
Fig. 1.
Work-flow overview. a. Experimental data acquisition from Aldrin-exposed and non-exposed DU145 prostate cancer cells. Relevant peak areas from non-targeted metabolomic an lipidomic experiments are set by applying MCR-ALS and ROI methods, next significant consumption/productions are determined through Mann-whitney test b. Algorithm-based automatic gene-to-reaction association building. The algorithm uses a variety of data bases to generate a set of gene-to-protein associations for each reaction with enzymatic activity in a model. c. transcriptomic data integration via either GPR and S-GPR into a GSMM reconstruction analysis by applying four different constraint-based methods d. model prediction of metabolic consumption/production e. Validation of the prediction by comparing predicted and experimental metabolic consumption/production (Fisher exact test) f. Evaluate the improvement in model predictions provided by the incorporation of stoichiometry into the gene-to-reaction associations (S-GPR)