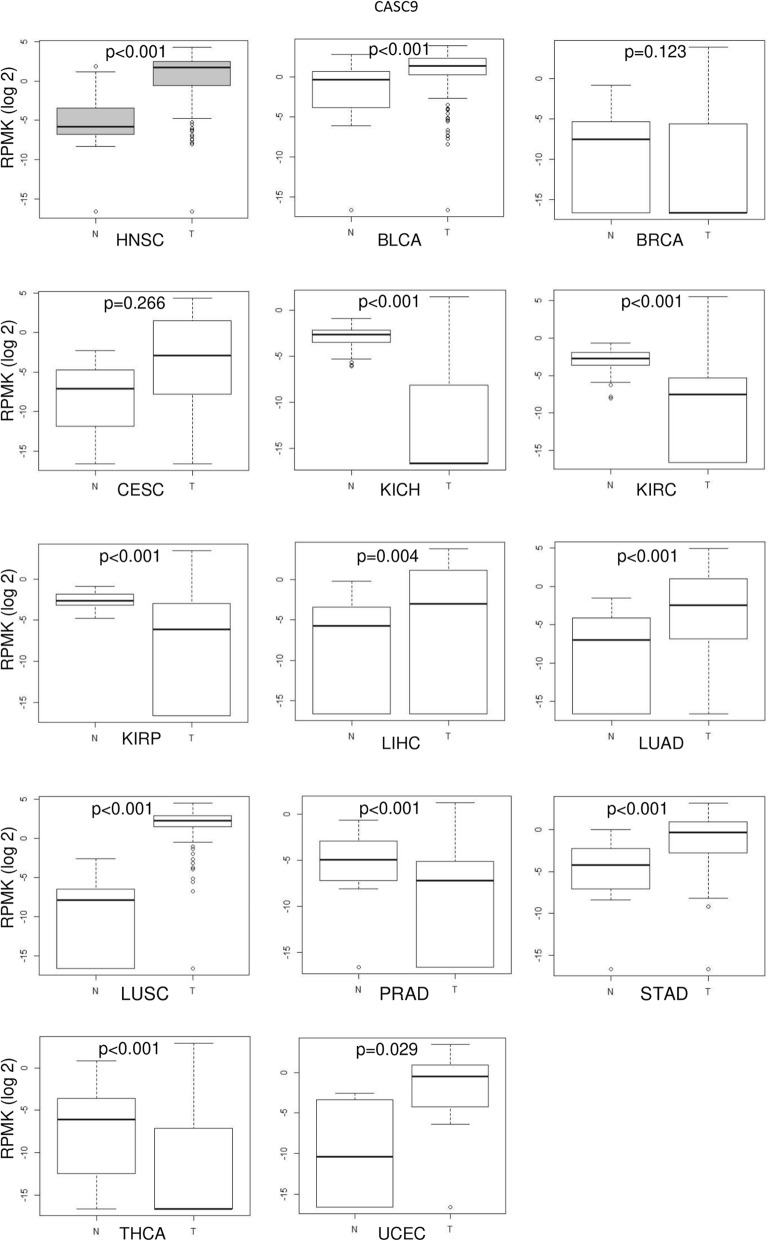
Fig. 3.
Pan-cancer analysis of CASC9 expression in TCGA datasets. The TANRIC database was used to access publicly available RNA-Seq data for CASC9 expression in various tumor entities: HNSC: head-neck squamous cell carcinoma; BLCA: urothelial bladder carcinoma; BRCA: breast invasive carcinoma; CESC: cervical squamous cell carcinoma and endocervical adenocarcinoma; KICH: kidney chromophobe; KIRC: kidney renal clear cell carcinoma; KIRP: kidney renal papillary cell carcinoma; LIHC: liver hepatocellular carcinoma; LUAD: lung adenocarcinoma; LUSC: lung squamous cell carcinoma.; PRAD: prostate adenocarcinoma; STAD: stomach adenocarcinoma; THCA: thyroid cancer; UCEC: uterine corpus endometrial carcinoma. LncRNA expression values were obtained as log2 RPMK (reads per kilo base per million mapped reads). Mann-Whitney U-test was applied to calculate p-values for differences between control (N) and tumour (T) samples