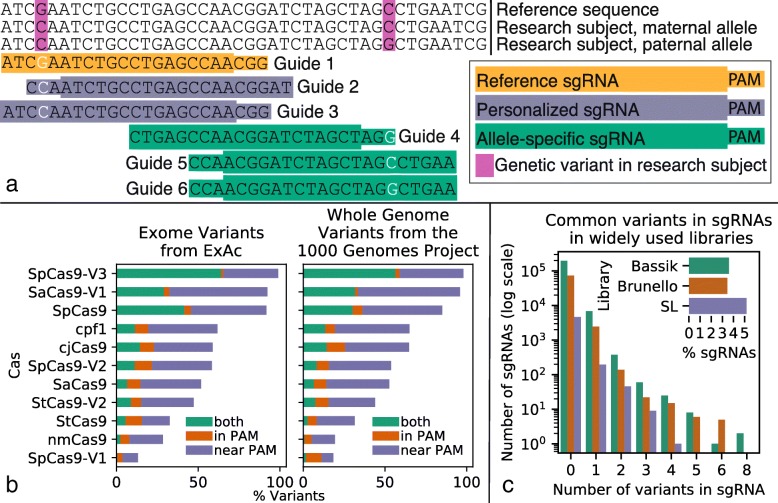
Fig. 1.
Analysis of allele-specific sgRNA sites. a In a sample genome, tools designing sgRNAs for the reference genome are imperfect matches due to genetic variants, exemplified by guide 1. AlleleAnalyzer designs personalized sgRNAs, as demonstrated by guides 2 and 3, which incorporate homozygous and avoid heterozygous variants, thus designing a guide perfectly matched to both alleles in a subject. It also designs personalized allele-specific sgRNAs based on the incorporation of heterozygous variants, shown by guides 4–6. Guides 4 and 6 target the paternal allele, while guide 5 targets the maternal allele. b Most variants annotated by the 1000 Genomes Project (1KGP) and the Exome Aggregation Consortium (ExAc) are in or near a PAM site. c Analysis of common variants (minor allele frequency (MAF) greater than 5% in 1KGP) and all variants in an individual cell line (WTC) within commonly used sgRNA libraries