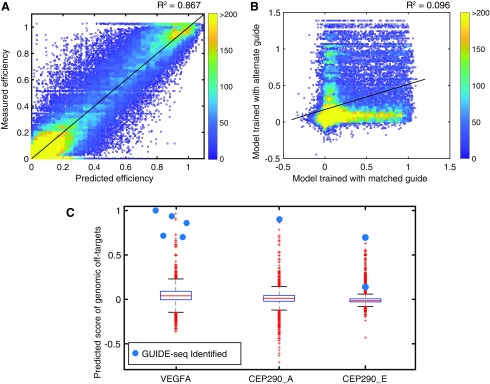
FIG. 4.
Guide-specific modeling predicts cleavage of off-target libraries in vitro. Neural network models were trained on data collected on reactions run for 30 min with 10:1 RNP:target library. (A) Predicted cutting efficiency versus measured efficiency of libraries of off-targets. Color indicates density of points. (B) Neural network models are highly guide specific. We applied either a prediction from a neural network model generated from a CEP290_C library or a prediction from a model generated from a VEGFA library, and applied these predictions to VEGFA off-targets. The results from each model are graphed. (C) Box plot of predicted efficiencies on genomic off-targets of Staphylococcus aureus complexed to VEGFA, CEP290_A, or CEP290_E (up to six mismatches). Sites identified by GUIDE-seq are indicated by larger blue dots.17