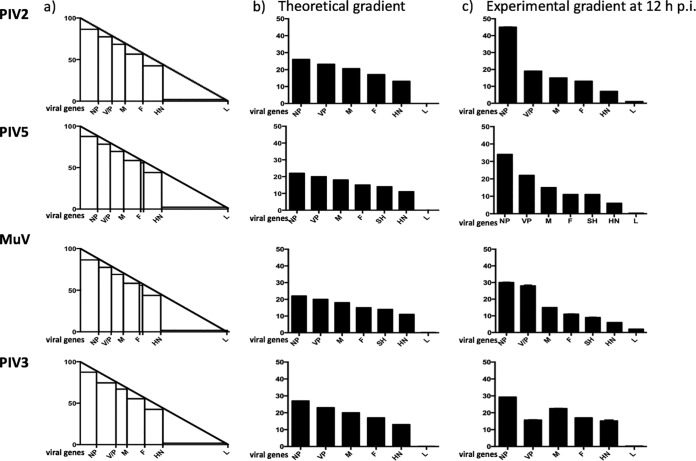
FIG 8.
Theoretical mRNA abundance gradients compared to actual gradients in a model in which vRdRP disengages with equal chance at any nucleotide during transcription, and truncated, non-poly(A) mRNAs are rapidly degraded. (a) Model of the relative abundance of individual viral mRNAs in which position 1 of the genome constitutes 100% of transcripts and the last nucleotide constitutes 1 to 2%. The end of each gene is indicated where polyadenylation occurs at the U tract to generate mRNAs that are subsequently translated. In this model it is assumed that transcripts that are prematurely terminated when vRdRP disengages from the genome upstream of the U tract are not polyadenylated and are degraded rapidly. The stepwise transcription profiles therefore reflect the theoretical abundance of polyadenylated mRNAs. (b) The theoretical percentage contribution of polyadenylated viral mRNAs to the total viral mRNA population, as calculated from the theoretical gradient shown in panel a. (c) The mRNA abundance gradient determined experimentally for cells infected with PIV2-Co, PIV5-W3, MuV, or PIV3-Wash at 12 h p.i. as described for Fig. 2.