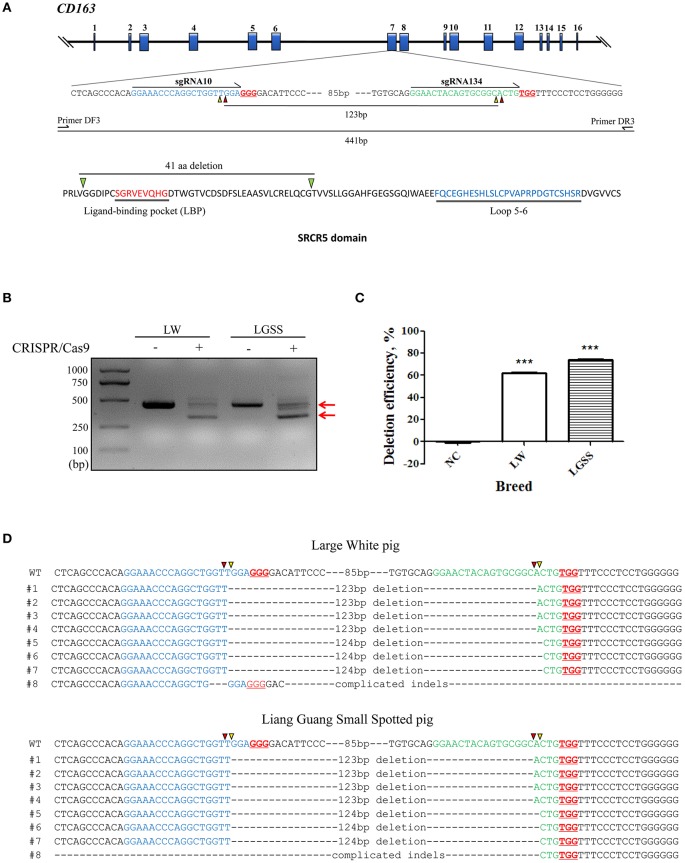
Figure 1.
Generation of the precise partial deletion of CD163 SRCR5 in porcine embryonic fibroblasts (PEFs) using CRISPR/Cas9. (A) Schematic of the CD163 gene and target sites of sgRNAs designed for targeting SRCR5 in the exon 7. The 16 exons of CD163 are indicated by blue rectangles. Arrows indicate the sequence used for the guide segment of sgRNA10 and 134. The NGG nucleotide protospacer adjacent motif sequences are underlined in red. Red and yellow triangles represent the predicted cleavage sites of sgRNAs. A precise excision with paired sgRNAs results in a 123 bp in-frame deletion including ligand-binding pocket (LBP). The primer pair DF3/DR3 was used to amplify a 441 bp product from the intact allele of CD163 gene and a truncated product of 317 bp if the deletion (123 bp) has occurred. Two regions (LBP and loop 5–6) of SRCR5 are shown. (B) PCR products identifying the presence of the targeted deletion of CD163 SRCR5 induced by paired sgRNAs. The upper red arrow indicates the position of the 441 bp full length PCR product, and the lower red arrow indicates the expected positions of the truncated PCR product in the event of deletion. LW, Large White pig; LGSS, Liang Guang Small Spotted pig; M, marker. (C) The efficiency of the targeted deletion in PEFs was quantified by qPCR. ***p < 0.001 compared to negative control. (D) Sequence analysis of cloned PCR products. The guide segments of sgRNA 10 and 134 are shown in blue and green, respectively. Red and yellow triangles represent the predicted cleavage sites of sgRNAs. WT, wild-type DNA sequence. Data are representative of the results of three independent experiments (means ± SE). Significant differences are indicated as follows: ***p < 0.001.