Abstract
Early onset breast cancer is a common malignancy and cause of death among young women in KSA. In addition, the data from women have demonstrated that most patients present late with an advanced stage. The early detection of this disease would not only save patients' lives but would also have the potential to reduce the budget and the time required for treating and nursing advanced breast cancer patients. This review highlights the risk of developing breast cancer in women with the methylated BRCA1 promoter in their white blood cells and proposes the potential use of this epigenetic modification as a powerful molecular marker for the early detection of breast cancer.
Keywords: BRCA1, Breast cancer, Epigenetic, Epigenetic modification, Methylation
الملخص
سرطان الثدي المبكر ورم خبيث منتشر، وهو أحد أسباب الوفاة بين النساء الشابات في المملكة العربية السعودية. إلى جانب ذلك، أظهرت البيانات أن غالبية المرضي يتقدمون للعلاج في مراحل المرض المتقدمة. والكشف المبكر لهذا المرض لا ينقذ حياة المرضى فقط، ولكن يمكن أن يخفض من المال والوقت المطلوبين لعلاج وتمريض مرضى سرطان الثدي المتقدم. يسلط هذا الاستعراض الضوء على خطر الإصابة بسرطان الثدي لدى النساء ممن يحملن مروج الجين رقم ١ المثبط لسرطان الثدي المُمَثْيَل في خلايا دمائهن البيضاء، ويقترح إمكانية استخدام هذا التعديل في التخلق المتوالي كعلامة جزيئية قوية للكشف المبكر عن سرطان الثدي.
الكلمات المفتاحية: التخلق المتوالي, مَثْيَلَة, الجين رقم١ المثبط لسرطان الثدي, سرطان الثدي, تعديل التخلق المتوالي
Introduction
Breast cancer among Arab women, as elsewhere in the world, is a common malignancy and cause of death, and its incidence is increasing. In KSA, 26.4% of all female breast cancers develop before the age of 40 compared to 6.5% in the USA. The breast cancer susceptibility gene, BRCA1, was discovered in 1994 as the first major gene associated with breast cancer.1 The hereditary type of breast cancer has been found to be attributed to germline mutations in BRCA1. These mutations account for approximately 5–10% of all breast cancers.2, 3 Furthermore, DNA methylation is the mechanism by which BRCA1 is inactivated during sporadic carcinogenesis.4 Both types of tumours occur at an early age and exhibit poor histological differentiation, Oestrogen and Progesterone receptor negativity and similar global gene expression profiles.5
The detection of the methylated BRCA1 promoter in DNA from peripheral blood and tumour tissues in breast cancer patients6 has suggested the involvement of this epigenetic modification, which occurs in normal non-epithelial tissue, in the development of breast cancer with BRCA1-like characteristics. However, it is still undetermined whether women carrying the methylated BRCA1 promoter in their WBC are at a high risk of breast cancer predisposition.
In this review, we explore the possible implication of BRCA1 promoter methylation in the development of breast cancer and propose the potential use of this aberrant methylation as a powerful non-invasive molecular marker for detecting predisposed individuals at an early age.
Breast cancer susceptibility gene: BRCA1
The human BRCA1 gene is a tumour suppressor gene that is located on the long (q) arm of chromosome 17. BRCA1 is expressed in cells in the breast and other tissues. BRCA1 plays a crucial role in the process of DNA repair, the control of cell cycle checkpoints and transcription. The loss of BRCA1 activity leads to tumour formation in specific target tissues. As BRCA1 is involved in the potentially error-free pathway of homologous recombination,7 which repairs double-strand breaks, cells that lack the BRCA1 protein tend to repair DNA damage by alternative error-prone mechanisms. This results in the generation of mutations and gross chromosomal rearrangements that can lead to carcinogenesis.7 Hence, females carrying germline BRCA1 pathogenic mutations are at an increased risk of developing aggressive breast and ovarian tumours characterized by poor histologic differentiation, high grade, aneuploidy, and hormone receptor negativity at an early age (<50).8
DNA methylation is an alternative mechanism for BRCA1 inactivation
Both BRCA1 mRNA and protein levels were found to be under-expressed in a subset of sporadic human breast cancers.9 These sporadic early onset breast cancers have aggressive pathologic features that are similar to those observed with mutated BRCA1. This finding suggested that, in the nonhereditary forms of breast cancer, alterations in BRCA1 or BRCA1-related pathway(s) might also play a role in the aggressiveness and pathogenesis of sporadic breast cancer. As no somatic mutations in BRCA1 were detected in the sporadic form of breast cancer, it was suggested that an epigenetic mechanism might be an alternative means by which BRCA1 is inactivated during this form of breast carcinogenesis.4 Indeed, the results from several studies revealed that 9–44% of sporadic breast cancer samples harboured methylated BRCA1 promoter.4, 10, 11
Structure of the 5′ regulatory region of BRCA1
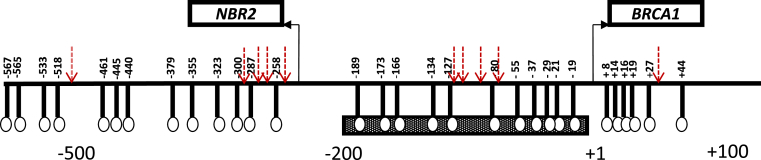
The 5′ regulatory promoter region of BRCA1 has been shown to contain 30 CpG sites overlying the area from −567 to +44 relative to the exon 1A transcription start site (Figure 1). A bi-directional core promoter (−218 to +1), which is located within this region, has been found to regulate the transcription of both the BRCA1 and the NBR2 genes.12 This 218 bp region is a CpG-rich area containing 11 CpG sites with a strong promoter activity13 that has been shown to be aberrantly hyper-methylated in human breast cancer cells and tissues14, 15, 16, 17 but not in normal human mammary epithelial cells.18
Figure 1.
Schematic representation of the BRCA1 promoter region. The middle hatched box represents the region of the BRCA1 core promoter. The bent arrows show the transcription start sites and directions. The vertical lines with circles indicate the positions of the 30 CpG sites. The numbers refer to the nucleotide positions relative to the BRCA1 transcription start. The red arrows indicate the position of methylation-related newly formed CpG sites.
Methylated BRCA1 promoter in peripheral blood DNA from breast cancer female patients
In 2008, Snell et al. have demonstrated the presence of the methylated BRCA1 promoter in normal non-epithelial tissues in patients from breast-ovarian cancer families.6 This finding suggested that the methylated BRCA1 promoter occurring in this tissue of the body is linked with BRCA1-like breast cancer development.6 This led the author to hypothesize that the deactivation of BRCA1 by promoter hyper-methylation might occur as a germline or an early somatic event, leading to breast cancer predisposition with a phenotype that is similar to that linked with BRCA1 germline mutations. Subsequent to Snell's study, several investigators have reported the detection of methylated BRCA1 in very young breast cancer patients,19, 20, 21, 22, 23 suggesting the potential use of methylated BRCA1 as a predictor of cancer risk.
In 2011, we have reported that 27.6% of primary sporadic breast carcinomas in Arab women comprise the hyper-methylated BRCA1 promoter.11 This occurrence is in the higher end of previously reported incidences of 7–44%.4, 10 Notably, the methylation of the BRCA1 promoter was found to be strongly associated with an early age onset of ≤40 years and is more common in high-grade tumours. Subsequently, in 2014, we have reported that 14.2% of breast cancer patients harboured the methylated BRCA1 promoter in their WBC. This was also significantly associated with the early onset of the disease.24 A high proportion of those patients (66.7%) exhibited methylated BRCA1 in matching tumour DNA. This result suggests that the presence of BRCA1 promoter methylation in WBC may elicit the development of breast cancer. Certainly, it has been postulated that constitutional BRCA1 promoter methylation may represent the “first-hit” predisposing and initiating tumourigenesis with morphologic features similar to those associated with BRCA1 germline mutations.25
Methylated BRCA1 promoter in peripheral blood DNA from cancer-free women
Snell et al. were the first to observe the presence of BRCA1 methylation in WBC DNA from a healthy female.6 This result led to the question of whether this female has a high risk of breast cancer predisposition in the future. Subsequently, several studies have reported the detection of methylated BRCA1 in WBC from normal healthy individuals.11, 19, 20, 21, 22, 23 We also have shown the presence of the methylated BRCA1 promoter in WBC of 9.7% of healthy cancer-free women (carriers). The majority of those carriers are ≤40 years old,11, 24 and 77% of them have cancer family histories, including breast and/or ovarian cancer.
Detection of methylation-related mutations throughout the BRCA1 promoter CpG Island
The use of high-resolution sodium bisulfite genomic sequencing of the BRCA1 promoter region has shown the presence of methylation-related mutations in WBC DNA from carriers and breast cancer patients.11 These types of mutations involve an association between cytosine methylation and T >C transitions, leading to the formation of novel CpG methylated sites. A number of these methylation-related mutations were found throughout the entire CpG Island, including the BRCA1 core promoter region (Figure 1). Although the functional significance of these mutations remains unknown, these mutations contribute to the overall methylation of the BRCA1 promoter region, suggesting their possible involvement in carcinogenesis. Indeed, several methylation-related mutations in the TP53 gene, which included those leading to the formation of new CpG sites, were found to predominate during lung carcinogenesis. Recently, the origin of T>C transition mutations in breast cancer has been revealed. It has been shown that these transition mutations are caused by DNA damage induced by Nitric Oxide, which is synthesized by the enzyme Nitric Oxide Synthase. This enzyme is enhanced in certain inflammatory environments and by oestrogen, and it is found to be over-expressed in the normal tissue adjacent to breast cancer.26 DNA damage caused by nitric oxide leads to the deamination of adenine to form hypoxanthine, which is then excised by the thymine DNA glycosylase base excision repair enzyme and repaired to C, resulting in the T >C transition. The majority of these mutations are observed in histologically normal tissues adjacent to breast cancer, and they occur most frequently in the 5′-ATG-3′, 5′-CTG-3′, and 5′-ATA-3′, sites.27
Methylated BRCA1 promoter in peripheral blood DNA and the risk of breast cancer predisposition
The following is an important question that still awaits a definite answer: Are carriers of the methylated BRCA1 promoter at a high risk of breast cancer predisposition? To answer this question, we hypothesized that if BRCA1 methylation in WBC presents a high risk of breast cancer predisposition, WBC from carriers should demonstrate molecular changes that are comparable, to some extent, to those identified in BRCA1-methylated WBC from breast cancer patients.24 Interestingly, we have demonstrated that cancer-free females harbouring the methylated BRCA1 promoter in their WBC have several breast cancer-related molecular changes that may provoke their potential predisposition for the development of breast cancer. We have reported that nine different breast cancer-related genes, in addition to BRCA1, were found to be epigenetically modified in WBC from both breast cancer patients and carriers. These genes are involved in various aspects of breast carcinogenesis, including tumour suppression (HIC1,28 CDH129, CDH1330, CDKN231), DNA repair (MGMT32), apoptosis (PYCARD,33 TNFRSF10C34), and cell cycle regulation (CCNA135). Furthermore, we have also reported that fifteen cancer-related genes in addition to BRCA1 were found to be differently expressed in the WBC from breast cancer patients and carriers. Two of these genes, ATM and insulin-like growth factor receptor (IGF1R), were found to be highly expressed in the WBC from carriers compared to that from the breast cancer cases. An elevation in the expression of either of these genes has been reported to be associated with an increase in the risk of future breast cancer.36, 37 We have also investigated the signature of plasma proteins in the carriers group and compared it with those in breast cancer patients and controls. In total, 35 proteins were found to be differentially expressed in the plasma from breast cancer patients, carriers, and controls. One of these proteins is Apolipoprotein CIII, which has been found to be down regulated in the plasma from pancreatic patients compared to that from controls.38, 39 Hence, this protein was reported to be a potential marker for the early detection of pancreatic cancer. Intriguingly, we have reported the down regulation of Apolipoprotein CIII to be 3- and 1.5-fold in plasma from breast cancer patients and carriers compared to controls, respectively. Altogether, these findings suggest the existence of a robust correlation between the methylated BRCA1 promoter in WBC and breast cancer-related molecular changes. Accordingly, these findings may infer that women carrying the methylated BRCA1 promoter in their peripheral blood DNA are at a high risk of breast cancer predisposition.
Conclusions
BRCA1 promoter methylation occurring in WBC appears to be linked with a high risk of BRCA1-like breast cancer development. The high prevalence of this epigenetic modification in WBC DNA of cancer-free women may contribute to the high proportion of early onset breast cancer in women in KSA. Recently, a meta-analysis involving 40 studies, including our 2011 study,11 was performed to obtain a more precise estimate of the association between BRCA1 methylation and sporadic breast cancer.40 The study indicated that BRCA1 promoter methylation emerged as a useful predictive biomarker for breast cancer in clinical assessments. This strongly suggests the potential use of BRCA1 promoter methylation in WBC as a molecular biomarker for the early prediction of breast cancer predisposition.
Author's contribution
NM is the sole author who conceived the idea of this review, revised the literature, wrote the initial draft, revised and edited the second draft. NM proof read the article and approved the final draft. NM is solely responsible for the content and the similarity index of this article.
Conflict of interest
The author has no conflict of interest to declare.
Acknowledgements
I would like to acknowledge Dr. Abdelilah Aboussekhra for proofreading the review.
Footnotes
Peer review under responsibility of Taibah University.
References
- 1.Bell R., Rosenthal J., Hussey C., Tran T., McClure M., Frye C., Hattier T., Phelps R., Haugen-Strano A., Katcher H., Yakumo K., Gholami Z., Shaffer D., Stone S., Bayer S., Wray C., Bogden R. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 2.Balmana J., Diez O., Rubio I.T., Cardoso F., Group E.G.W. BRCA in breast cancer: ESMO clinical practice guidelines. Ann Oncol Off J Eur Soc Med Oncol/ESMO. 2011;22(Suppl 6) doi: 10.1093/annonc/mdr373. vi31–4. [DOI] [PubMed] [Google Scholar]
- 3.Campeau P.M., Foulkes W.D., Tischkowitz M.D. Hereditary breast cancer: new genetic developments, new therapeutic avenues. Hum Genet. 2008;124:31–42. doi: 10.1007/s00439-008-0529-1. [DOI] [PubMed] [Google Scholar]
- 4.Birgisdottir V., Stefansson O.A., Bodvarsdottir S.K., Hilmarsdottir H., Jonasson J.G., Eyfjord J.E. Epigenetic silencing and deletion of the BRCA1 gene in sporadic breast cancer. Breast Cancer Res: BCR. 2006;8:R38. doi: 10.1186/bcr1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hedenfalk I., Duggan D., Chen Y., Radmacher M., Bittner M., Simon R., Meltzer P., Gusterson B., Esteller M., Raffeld M., Yakhini Z., Ben-Dor A., Dougherty E., Kononen J., Bubendorf L., Fehrle W., Pittaluga S., Gruvberger S., Loman N., Johannsson O., Olsson H., Wilfond B., Sauter G., Kallioniemi O.-P., Borg Å., Trent J. Gene-expression profiles in hereditary breast cancer. N. Engl J Med. 2001;344:539–548. doi: 10.1056/NEJM200102223440801. [DOI] [PubMed] [Google Scholar]
- 6.Snell C., Krypuy M., Wong E.M., kConFab i, Loughrey M.B., Dobrovic A. BRCA1 promoter methylation in peripheral blood DNA of mutation negative familial breast cancer patients with a BRCA1 tumour phenotype. Breast Cancer Res BCR. 2008;10:R12. doi: 10.1186/bcr1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacinto F.V., Esteller M. Mutator pathways unleashed by epigenetic silencing in human cancer. Mutagenesis. 2007;22:247–253. doi: 10.1093/mutage/gem009. [DOI] [PubMed] [Google Scholar]
- 8.Arver B., Du Q., Chen J., Luo L., Lindblom A. Hereditary breast cancer: a review. Seminars Cancer Biol. 2000;10:271–288. doi: 10.1006/scbi.2000.0325. [DOI] [PubMed] [Google Scholar]
- 9.Thompson M.E., Jensen R.A., Obermiller P.S., Page D.L., Holt J.T. Decreased expression of BRCA1 accelerates growth and is often present during sporadic breast cancer progression. Nat Genet. 1995;9:444–450. doi: 10.1038/ng0495-444. [DOI] [PubMed] [Google Scholar]
- 10.Butcher D.T., Rodenhiser D.I. Epigenetic inactivation of BRCA1 is associated with aberrant expression of CTCF and DNA methyltransferase (DNMT3B) in some sporadic breast tumours. Eur J Cancer. 2007;43:210–219. doi: 10.1016/j.ejca.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 11.Al-Moghrabi N., Al-Qasem A.J., Aboussekhra A. Methylation-related mutations in the BRCA1 promoter in peripheral blood cells from cancer-free women. Int J Oncol. 2011;39:129–135. doi: 10.3892/ijo.2011.1021. [DOI] [PubMed] [Google Scholar]
- 12.Xu C.F., Brown M.A., Nicolai H., Chambers J.A., Griffiths B.L., Solomon E. Isolation and characterisation of the NBR2 gene which lies head to head with the human BRCA1 gene. Hum Mol Genet. 1997;6:1057–1062. doi: 10.1093/hmg/6.7.1057. [DOI] [PubMed] [Google Scholar]
- 13.Xu CF, Chambers JA, Solomon E. Complex regulation of the BRCA1 gene. J Biol Chem 1997; 272:20994-7. [DOI] [PubMed]
- 14.Dobrovic A., Simpfendorfer D. Methylation of the BRCA1 gene in sporadic breast cancer. Cancer Res. 1997;57:3347–3350. [PubMed] [Google Scholar]
- 15.Mancini D.N., Rodenhiser D.I., Ainsworth P.J., O'Malley F.P., Singh S.M., Xing W., Archer T.K. CpG methylation within the 5' regulatory region of the BRCA1 gene is tumor specific and includes a putative CREB binding site. Oncogene. 1998;16:1161–1169. doi: 10.1038/sj.onc.1201630. [DOI] [PubMed] [Google Scholar]
- 16.Catteau A., Harris W.H., Xu C.F., Solomon E. Methylation of the BRCA1 promoter region in sporadic breast and ovarian cancer: correlation with disease characteristics. Oncogene. 1999;18:1957–1965. doi: 10.1038/sj.onc.1202509. [DOI] [PubMed] [Google Scholar]
- 17.Bianco T., Chenevix-Trench G., Walsh D.C., Cooper J.E., Dobrovic A. Tumour-specific distribution of BRCA1 promoter region methylation supports a pathogenetic role in breast and ovarian cancer. Carcinogenesis. 2000;21:147–151. doi: 10.1093/carcin/21.2.147. [DOI] [PubMed] [Google Scholar]
- 18.Rice J.C., Massey-Brown K.S., Futscher B.W. Aberrant methylation of the BRCA1 CpG island promoter is associated with decreased BRCA1 mRNA in sporadic breast cancer cells. Oncogene. 1998;17:1807–1812. doi: 10.1038/sj.onc.1202086. [DOI] [PubMed] [Google Scholar]
- 19.Gupta S., Jaworska-Bieniek K., Narod S.A., Lubinski J., Wojdacz T.K., Jakubowska A. Methylation of the BRCA1 promoter in peripheral blood DNA is associated with triple-negative and medullary breast cancer. Breast Cancer Res Treat. 2014;148:615–622. doi: 10.1007/s10549-014-3179-0. [DOI] [PubMed] [Google Scholar]
- 20.Bosviel R., Garcia S., Lavediaux G., Michard E., Dravers M., Kwiatkowski F., Bignon Y.J., Bernard-Gallon D.J. BRCA1 promoter methylation in peripheral blood DNA was identified in sporadic breast cancer and controls. Cancer Epidemiol. 2012;36 doi: 10.1016/j.canep.2012.02.001. e177–82. [DOI] [PubMed] [Google Scholar]
- 21.Bosviel R., Michard E., Lavediaux G., Kwiatkowski F., Bignon Y.J., Bernard-Gallon D.J. Peripheral blood DNA methylation detected in the BRCA1 or BRCA2 promoter for sporadic ovarian cancer patients and controls. Clin Chimica Acta Int J Clin Chem. 2011;412:1472–1475. doi: 10.1016/j.cca.2011.04.027. [DOI] [PubMed] [Google Scholar]
- 22.Iwamoto T., Yamamoto N., Taguchi T., Tamaki Y., Noguchi S. BRCA1 promoter methylation in peripheral blood cells is associated with increased risk of breast cancer with BRCA1 promoter methylation. Breast cancer Res Treat. 2011;129:69–77. doi: 10.1007/s10549-010-1188-1. [DOI] [PubMed] [Google Scholar]
- 23.Cho Y.H., Yazici H., Wu H.C., Terry M.B., Gonzalez K., Qu M., Dalay N., Santella R.M. Aberrant promoter hypermethylation and genomic hypomethylation in tumor, adjacent normal tissues and blood from breast cancer patients. Anticancer Res. 2010;30:2489–2496. [PMC free article] [PubMed] [Google Scholar]
- 24.Al-Moghrabi N., Nofel A., Al-Yousef N., Madkhali S., Bin Amer S.M., Alaiya A., Shinwari Z., Al-Tweigeri T., Karakas B., Tulbah A., Aboussekhra A. The molecular significance of methylated BRCA1 promoter in white blood cells of cancer-free females. BMC Cancer. 2014;14:830. doi: 10.1186/1471-2407-14-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong E.M., Southey M.C., Fox S.B., Brown M.A., Dowty J.G., Jenkins M.A., Giles G.G., Hopper J.L., Dobrovic A. Constitutional methylation of the BRCA1 promoter is specifically associated with BRCA1 mutation-associated pathology in early-onset breast cancer. Cancer Prev Res. 2011;4:23–33. doi: 10.1158/1940-6207.CAPR-10-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi M.R.M., Brown C., Clare S.E. Abstract P6-07-11: on the origin of T>C transition mutations in breast cancer. Cancer Res. 2016:76. [Google Scholar]
- 27.McGranahan N., Favero F., de Bruin E.C., Birkbak N.J., Szallasi Z., Swanton C. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci Transl Med. 2015;7 doi: 10.1126/scitranslmed.aaa1408. 283ra54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wales M.M., Biel M.A., el Deiry W., Nelkin B.D., Issa J.P., Cavenee W.K., Kuerbitz S.J., Baylin S.B. p53 activates expression of HIC-1, a new candidate tumour suppressor gene on 17p13.3. Nat Med. 1995;1:570–577. doi: 10.1038/nm0695-570. [DOI] [PubMed] [Google Scholar]
- 29.Guilford P.J., Hopkins J.B.W., Grady W.M., Markowitz S.D., Willis J., Lynch H., Rajput A., Wiesner G.L., Lindor N.M., Burgart L.J., Toro T.T., Lee D., Limacher J.-M., Shaw D.W., Findlay M.P.N., Reeve A.E. E-cadherin germline mutations define an inherited cancer syndrome dominated by diffuse gastric cancer. Hum Mutat. 1999;14:249–255. doi: 10.1002/(SICI)1098-1004(1999)14:3<249::AID-HUMU8>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 30.Lee S.W. H-cadherin, a novel cadherin with growth inhibitory functions and diminished expression in human breast cancer. Nat Med. 1996;2:776–782. doi: 10.1038/nm0796-776. [DOI] [PubMed] [Google Scholar]
- 31.Lukas J., Parry D., Aagaard L., Mann D.J., Bartkova J., Strauss M., Peters G., Bartek J. Retinoblastoma-protein-dependent cell-cycle inhibition by the tumour suppressor p16. Nature. 1995;375:503–506. doi: 10.1038/375503a0. [DOI] [PubMed] [Google Scholar]
- 32.Pegg A.E. Mammalian O6-alkylguanine-DNA alkyltransferase: regulation and importance in response to alkylating carcinogenic and therapeutic agents. Cancer Res. 1990;50:6119–6129. [PubMed] [Google Scholar]
- 33.Srinivasula S.M., Poyet J.L., Razmara M., Datta P., Zhang Z., Alnemri E.S. The PYRIN-CARD protein ASC is an activating adaptor for caspase-1. J Biol Chem. 2002;277:21119–21122. doi: 10.1074/jbc.C200179200. [DOI] [PubMed] [Google Scholar]
- 34.Ashkenazi A. Targeting death and decoy receptors of the tumour-necrosis factor superfamily. Nat Rev Cancer. 2002;2:420–430. doi: 10.1038/nrc821. [DOI] [PubMed] [Google Scholar]
- 35.Yang R., Muller C., Huynh V., Fung Y.K., Yee A.S., Koeffler H.P. Functions of cyclin A1 in the cell cycle and its interactions with transcription factor E2F-1 and the Rb family of proteins. Mol Cell Biol. 1999;19:2400–2407. doi: 10.1128/mcb.19.3.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamimi R.M., Colditz G.A., Wang Y., Collins L.C., Hu R., Rosner B., Irie H.Y., Connolly J.L., Schnitt S.J. Expression of IGF1R in normal breast tissue and subsequent risk of breast cancer. Breast cancer Res Treat. 2011;128:243–250. doi: 10.1007/s10549-010-1313-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Angele S., Hall J. The ATM gene and breast cancer: is it really a risk factor? Mutat Res. 2000;462:167–178. doi: 10.1016/s1383-5742(00)00034-x. [DOI] [PubMed] [Google Scholar]
- 38.Chen J., Anderson M., Misek D.E., Simeone D.M., Lubman D.M. Characterization of apolipoprotein and apolipoprotein precursors in pancreatic cancer serum samples via two-dimensional liquid chromatography and mass spectrometry. J Chromatogr A. 2007;1162:117–125. doi: 10.1016/j.chroma.2007.03.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Honda K., Okusaka T., Felix K., Nakamori S., Sat N., Nagai H., Ioka T., Tsuchida A., Shimahara T., Shimahara M., Yasunami Y., Kuwabara H., Sakuma T., Otsuka Y., Ota N., Shitashige M., Kosuge T., Bü chler M.W., Yamada T. Altered plasma apolipoprotein modifications in patients with pancreatic cancer: protein characterization and multi-institutional validation. PLoS One. 2012;7:e46908. doi: 10.1371/journal.pone.0046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L., Long X. Association of BRCA1 promoter methylation with sporadic breast cancers: evidence from 40 studies. Sci Rep. 2015;5:17869. doi: 10.1038/srep17869. [DOI] [PMC free article] [PubMed] [Google Scholar]