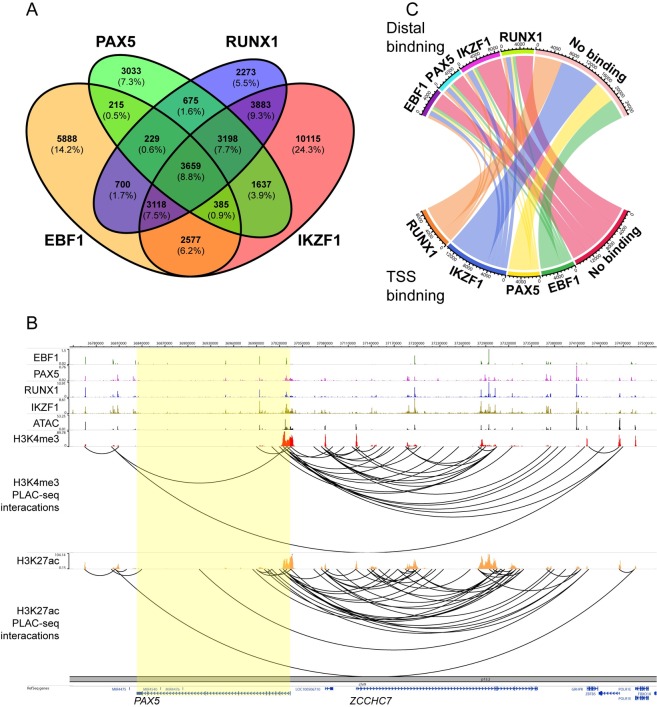
Fig 4. PAX5 is a transcriptional network key regulator in human leukemia cells.
Panel (A) display a 4-part Venn-diagram based on ChIP-seq experiments targeting PAX5, EBF1, RUNX1 and IKZF1 in the human PreB-ALL cell line NALM6. Overlapping ChIP-seq peaks were identified with the mergePeaks command in Homer. (B) Visualization of H3K4me3 and H3K27ac anchored PLAC-seq interactions at the PAX5 gene in NALM6 B-ALL cells. Binding of EBF1, PAX5, RUNX1 and IKZF1 as well as H3K4me3 and H3K27ac chromatin marks was determined by ChIP-seq. ATAC-seq was used to determine chromatin accessibility. The data was displayed using the WashU Genome Browser. (C) Chord diagram based on H3K4me3 anchored PLAC-seq from NALM6 displaying interactions between TSS and distal elements. PAX5-EBF1-RUNX1-IKZF1 gene networks anchored distal-to-TSS (transcription start site) are indicated in the diagram. Chromatin loops were filtered for overlapping PAX5, EBF1, RUNX1 or IKZF1 ChIP-seq peaks in either one or both anchor-points (for details, see Methods) with the constraints that one anchor-point had to be within 2.5kb from TSS and the other had to be located more than 2.5kb away from TSS (distal binding). The chord diagram was visualized with the circlize package in R.