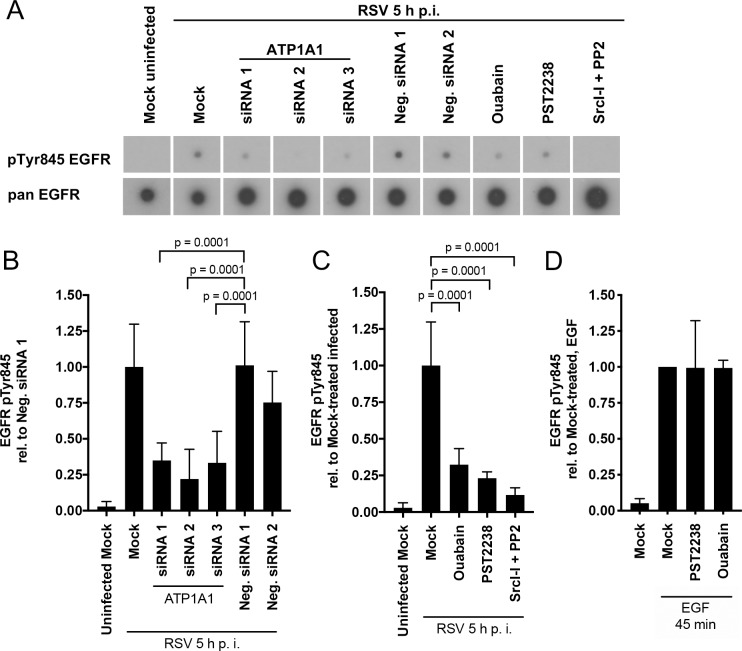
Fig 8. ATP1A1-dependent EGFR phosphorylation at Tyr845 during RSV infection.
A549 cells were treated as indicated (either siRNA knockdown for 48 h, or pre-treatment with the chemical compounds ouabain [25 nM], PST2238 [40 μM] or Src-Inhibitor-I (SrcI-I) [6.25 μM] and PP2 [12.5 μM] for 5 h pre-inoculation). Coincident with these treatments, the cells were serum starved for 16 h, and then were inoculated with wt RSV (MOI = 5 PFU/cell) and incubated for 5 h. The cells were lysed and lysates were incubated with a phospho-specific EGFR antibody array, followed by detection of bound EGFR with a biotinylated pan (i.e., universal) EGFR antibody, followed by incubation with horse radish peroxidase-conjugated streptavidin, and visualization with X-ray film (RayBiotech, Inc.). (A) X-ray film showing representative array spots of pTyr845 EGFR and its corresponding pan EGFR for each treatment. (B—D) Quantification of EGFR Tyr845 phosphorylation. pTyr845 EGFR signals of three independent experiments with two technical replicates each were quantified by scanning and normalized to the signal of the internal array positive controls and pan EGFR. (B) shows the siRNA knockdown samples reported relative to Neg. siRNA 1-transfected, RSV-infected samples, and (C) shows the chemical-treated samples reported relative to mock-treated, RSV-infected samples. (D) As control, PST2238 or Ouabain pretreated A549 cells were stimulated with EGF (100 ng/ml) for 45 min and the pTyr845 EGFR signal was quantified. The statistical significance of the differences were determined by one-way analysis of variance with Dunnett multiple-comparison test and the p-values are indicated for each comparison.