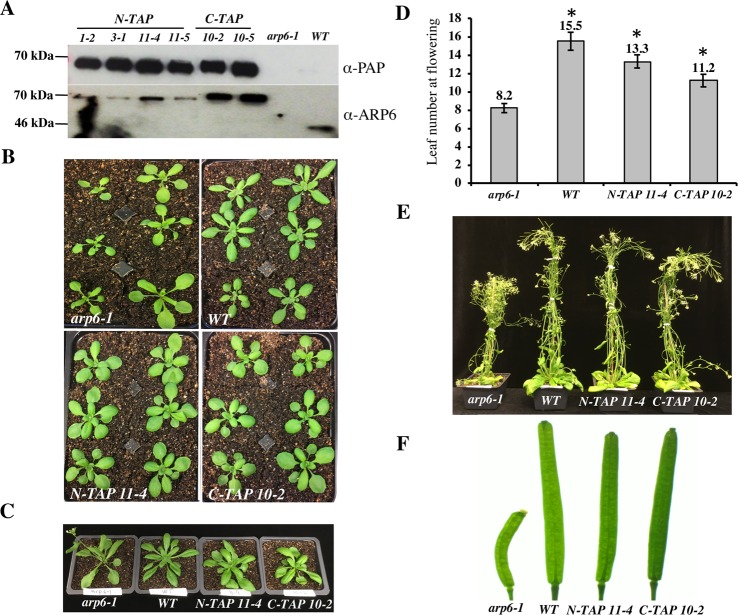
Fig 1. N-TAP-ARP6 and C-TAP-ARP6 transgenes rescue the arp6-1 phenotype.
(A- upper blot) T2 plants homozygous for N-TAP- or C-TAP-ARP6 transgenes express a fusion protein with the expected size of 67.5 kDa. The fusion protein is specifically detected only in transgenic plants and not in arp6-1 or WT plants using a peroxidase anti-peroxidase (PAP) soluble complex, which binds the protein A moiety of the TAP tag. (A- lower blot) The same protein extracts as in the upper blot were probed with a monoclonal ARP6 antibody. ARP6 presence is specifically detected in all transgenic plants as a 67.5 kDa fusion protein band compared to the 44 kDa ARP6 band in WT plants, and is absent in arp6-1 mutant plants. The ARP6 antibody reacts less strongly with N-TAP-ARP6, most likely because the antibody recognizes the N-terminal region, which is adjoined to the TAP tag in this fusion. (B) Transgenic plants look more similar to WT plants than arp6-1 plants, with more compacted, non-serrated rosette leaves. (C) Early flowering phenotype is rescued in transgenic plants when compared to the arp6-1. (D) The average number of rosette leaves of N-TAP-ARP6 and C-TAP-ARP6 transgenic plants at flowering is significantly higher than in arp6-1 (n = 6 for WT and arp6-1, and n = 12 for N-TAP 11–4 and C-TAP 10–2). Asterisks indicate significant differences from arp6-1 plants with p<0.001, calculated using unpaired t-tests. (E) The loss of apical dominance defects of arp6-1 plants are rescued in N-TAP-ARP6 and C-TAP-ARP6 transgenic plants. (F) The fertility defects of arp6-1 plants are rescued in N-TAP 11–4 and C-TAP 10–2 transgenic plants.