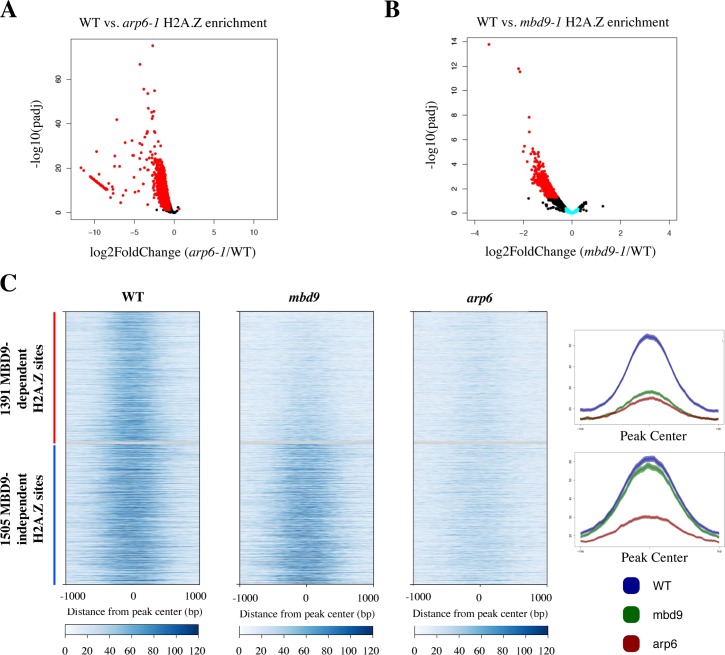
Fig 3. Identification of 1391 H2A.Z-enriched sites that require MBD9 for H2A.Z incorporation into chromatin.
(A-B) Volcano plots of–log10 of the adjusted P value (y-axis) versus log2 fold change of H2A.Z ChIP-seq reads (x-axis) between wild type plants and arp6-1 mutant plants (A) or mbd9-1 mutant plants (B). Each point corresponds to a called H2A.Z peak that was analyzed by DESeq2. Peaks that had a log2 fold change equal to or greater than 0.6 and an adjusted p value of 0.05 or less are colored red. Out of 7,039 peaks analyzed, there are 6,266 peaks that are significantly depleted of H2A.Z in arp6-1 plants (A, red dots), and 1,391 peaks that are significantly depleted of H2A.Z in mbd9-1 plants (MBD9-dependent peaks, panel B, red dots). Peaks that had an absolute log2 fold change from 0 to 0.25 in the WT to mbd9-1 comparison are colored light blue (MBD9-independent peaks, panel B, light blue dots). (C) Heatmaps (left) and average plots (right) of the 1391 MBD9-dependent (red bar, top of heatmaps, top average plot) peaks that are significantly depleted of H2A.Z in mbd9-1 plants and 1,505 MBD9-independent (blue bar, bottom of heatmaps, bottom average plot) peaks that had an absolute log2 fold change from 0 to 0.25 in the WT to mbd9-1 plants. Plots are centered on each peak and show a 2 kb window around the peak centers.