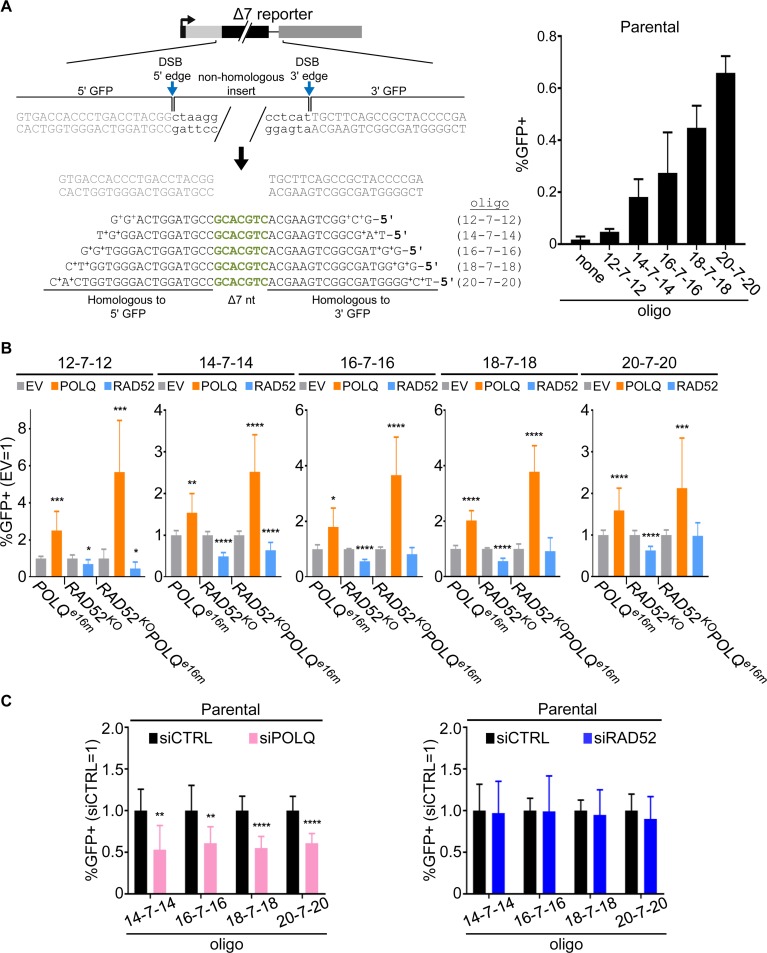
Fig 5. POLQ promotes oligonucleotide microhomology-templated repair events requiring nascent DNA synthesis.
(A) Diagram of Δ7 reporter in which 7 nt have been deleted from the 3' GFP sequence. The GFP coding sequence can be restored via templated insertion of the omitted 7 nt, which is provided by transfection of oligonucleotides. Five different oligonucleotides were used, which contain these 7 nt flanked on both sides by 12, 14, 16, 18, or 20 nt of homology to the 5' and 3' GFP sequences (12-7-12, 14-7-14, 16-7-16, 18-7-18, or 20-7-20, respectively). The plus signs indicate phosphorothioate linkages. These events are induced by expression of two sgRNAs and Cas9 to target DSBs at the edges of the 5' and 3' GFP sequences, thereby excising the non-homologous insert. These transfections also included the oligonucleotide templates and empty vector (EV). Shown are the percentages of GFP+ cells from these transfections, normalized to transfection efficiency, either in the absence (none) or presence of the 12-7-12, 14-7-14, 16-7-16, 18-7-18, or 20-7-20 oligonucleotides. Two independent clones were tested for each reporter with four independent replicates for a total n = 8, and error bars represent SD. (B) Influence of RAD52 and POLQ on repair events using oligonucleotide templates. RAD52KO, POLQe16m, and RAD52KOPOLQe16m Δ7 reporter cell lines were transfected as in (A), but including EV, RAD52 expression vector, or POLQ expression vector. GFP+ frequencies are normalized to transfection efficiency and shown relative to EV (EV = 1). Error bars represent SD. Two independent clones were tested for each reporter. n = 8 for 12-7-12, 14-7-14, 16-7-16, and n = 16 for 18-7-18 and 20-7-20. † P < 0.05 (unadjusted P-value), * P < 0.05, ** P < 0.01, *** P < 0.005, **** P < 0.001, EV vs. complementation, using unpaired t-test with Holm-Sidak correction. (C) The parental Δ7 reporter cell line was examined with siRNA treatments as in Fig 3. Two independent clones were tested for each treatment with four independent replicates for a total n = 8, and error bars represent SD. ** P < 0.01, **** P < 0.001, siCTRL vs. siRNA treatment using unpaired t-test with Holm-Sidak correction.