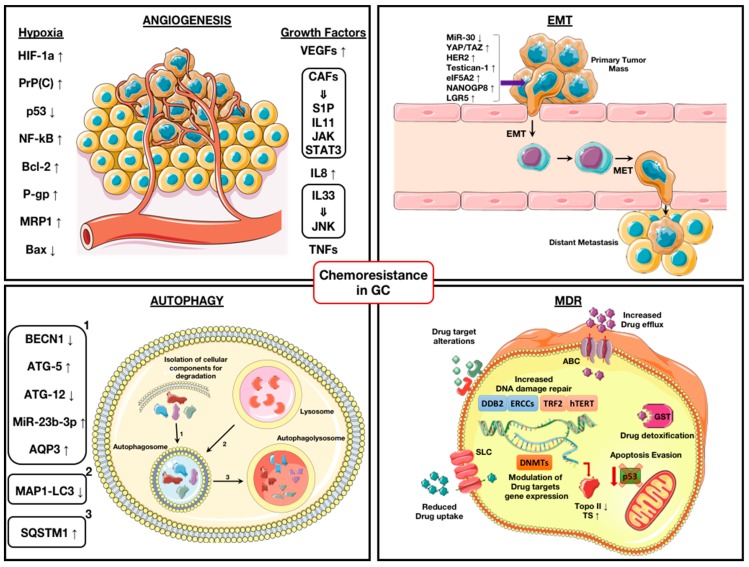
Figure 1.
Schematic representation of the most common mechanisms involved in chemo-resistance of gastric cancer (GC) cells. Angiogenesis: hypoxia regulated genes and growth factors released by GC and cancer-associated fibroblasts (CAFs) implicated in chemo-resistance. Epithelial-Mesenchymal Transition (EMT): factors involved in epithelial-mesenchymal switch that makes cancer cells drug-resistant and able to metastasize. Autophagy: proteins involved in (1) isolation of cellular components for degradation in autophagosome, (2) fusion between autophagosome and lysosome, and (3) catabolic processes in autophagolysosome and found associated with chemo-resistance. Multidrug Resistance (MDR): proteins involved in reduced drug uptake or increased drug efflux; increased drug detoxification; mutation or down-regulation of drug targets; up-regulation of drug targets; increased drug-induced DNA damage repair or telomere maintenance pathways; epigenetic regulation of gene expression. Abbreviations: solute carrier (SLC) and ATP-binding cassette (ABC) transporters; glutathione S-transferase (GST); Topo II (Topoisomerase II); thymidylate synthase (TS); DNA methyltransferases (DNMTs). (Images built with illustrations from https://smart.servier.com under CC 3.0 license).