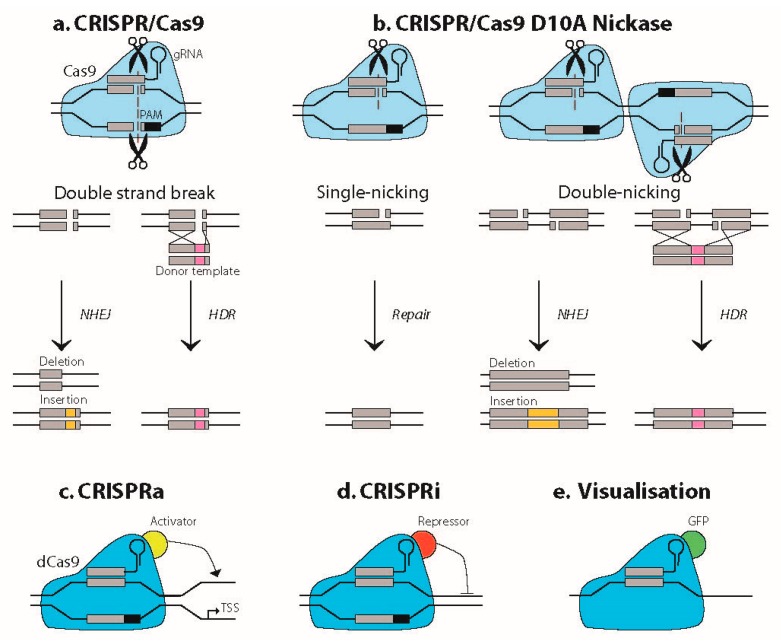
Figure 1.
Principles of main CRISPR/Cas9 applications. (a) CRISPR/Cas9 generates a double strand break (DSB) in the genome, guided by its small guide RNA (sgRNA) next to a proto-spacer adjacent motif (PAM) sequence. The DSB will in most cases be repaired by error-prone non-homologous end joining (NHEJ), resulting in the formation of an indel. Precise DNA repair will occur through homology-directed repair (HDR) and the use of a suitable donor template in a minority of the cases. (b) Cas9 D10A nickase (Cas9n) is mutated in one of its nuclease domains and will therefore introduce a single strand break in the DNA. When two nickases are targeted close to each other, the two nicks effectively generate a DSB, which will be followed by the same repair events as illustrated in a. (c–e) dCas9 is a double mutant and enzymatically inactive. It is being used as a precise and effective guiding vehicle to the genome and to transcripts. Examples shown here are transcription activation (CRISPRa) and transcription interference (CRISPRi) by dCas9 fusions to transcription activators and repressors, respectively. Fusion of GFP to dCas9 has been used to localize (CUG)n RNA in situ in living cells [6].