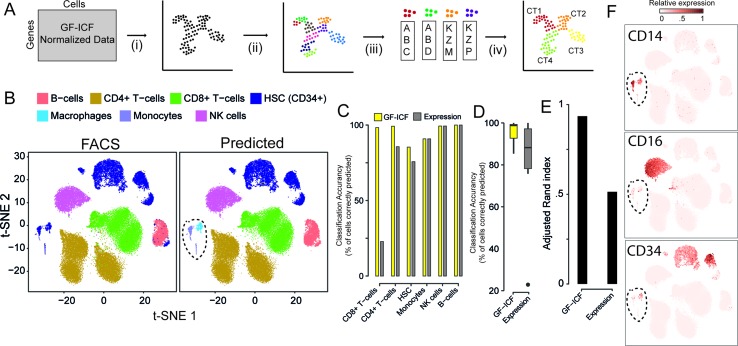
Figure 2.
Relevant genes extracted from the gf-icf pipeline enable cell type prediction. (A) Pipeline for the identification of cell type using gf-icf pipeline. Single-cell transcriptional profiles are normalized by gf-icf in order to score genes in each single-cell, and then (i) cells are projected with t-SNE in a bi-dimensional space; (ii) cells are divided in small groups using Louvain–Jaccard clustering; and (iii) the gene signature of each cluster is identified and (iv) used to predict cell type of origin by gene set enrichment analysis (GSEA) against a set bulk transcriptomic data of pure cell types. (B) Comparison between FACS-sorted cell type (left) and predicted cell type (right) of about 40k PBMCs. (C) Cell type prediction accuracy as a percentage of correctly predicted cells using either gf-icf or normalized counts. (D) Distribution of cell type prediction accuracy using either gf-icf or normalized counts. (E) Adjusted Rand index of cell type prediction using either gf-icf or normalized counts. (F) Expression of CD14, CD16, and CD34 marker genes for a small subpopulation of HSCs predicted instead to be monocyte and macrophages.