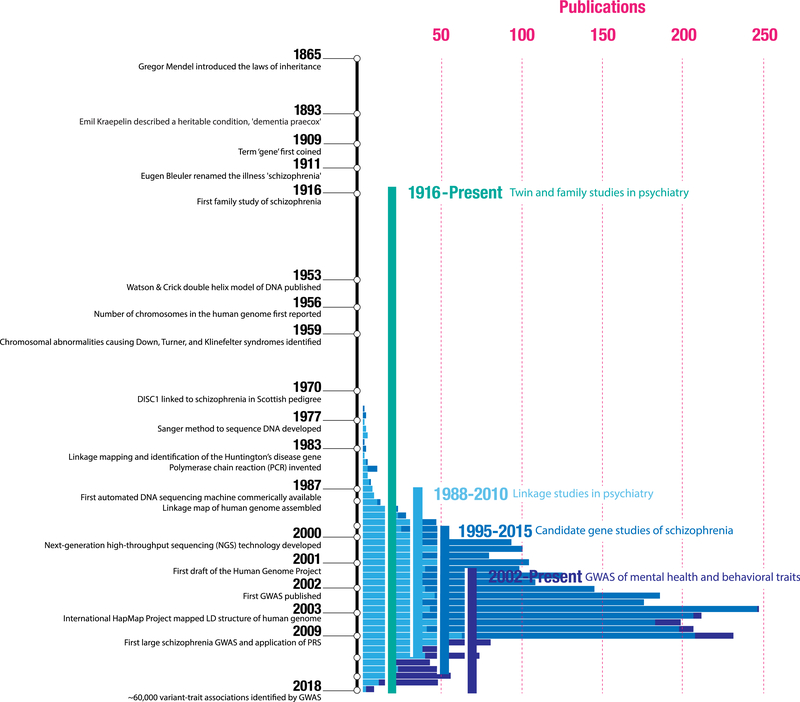
Figure 1.
Selected events from the methodological evolution of genetic analysis and, specifically, for psychiatric genetics, are shown on the vertical timeline (not to scale). The x-axis also illustrates a broad estimate of the number of articles per year for linkage studies in psychiatry (derived from PubMed), candidate gene studies in schizophrenia (SZGene; updated through 2012), and mental health and behavioral genome-wide association studies (GWASs) (from the National Human Genome Research Institute–European Bioinformatics Institute GWAS catalog). While the notion that schizophrenia was heritable was confirmed in the first half of the 20th century by twin and family studies, it only became possible to link genetic variation to psychiatric outcomes in the last 50 years. Cytogenetic studies, which detected large chromosomal abnormalities, were the first to investigate the genetic underpinnings of schizophrenia. Linkage studies scanned for patterns of disease cosegregation in large pedigrees (i.e., regions where a genetic divergence within a family consistently tracked with the individuals that developed the illness) to nominate broad loci that might harbor risk variants. Candidate gene studies moved genetic association outside of pedigrees to a case-control design. Here, the effect of a variant hypothesized to play a role in disease pathophysiology (e.g., variants within genes involved in dopaminergic signaling) on risk for an illness was assessed by comparing cases and controls. Finally, GWASs—a hypothesis-free approach where millions of variants were compared between cases and control subjects—became possible after the human genome was sequenced and catalogs of its structure were assembled (e.g., HapMap). Today, this is still the most popular approach to understanding how common genetic variation relates to disease. LD, linkage disequilibrium; PRS, polygenic risk score.