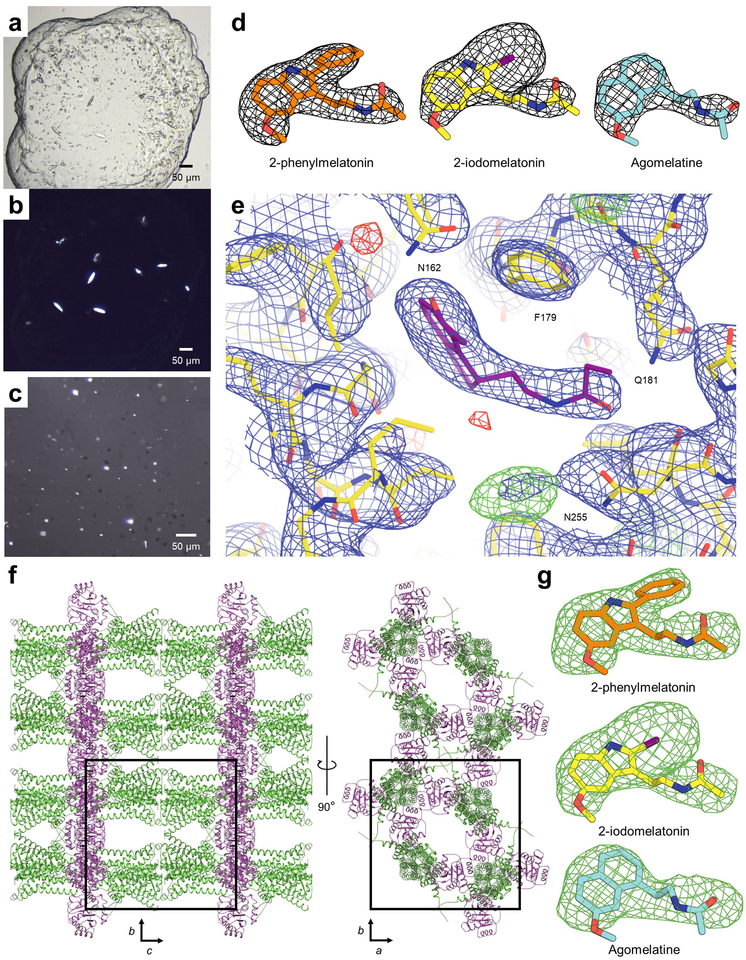
Extended Data Fig. 1 |. Crystals, ligand electron density maps, and packing of MT1.
a, Bright field, and b, cross-polarised images of representative MT1-2-pmt crystals, optimised for synchrotron data collection (representing three independent crystallization setups). c, Cross-polarised image of representative MT1-ramelteon crystals used for XFEL data collection (representing two independent crystallization setups). d, 2mFo−DFc ligand electron density maps of MT1 co-crystallised with 2-pmt (orange), 2-iodomelatonin (yellow), and agomelatine (cyan), contoured at 1.0 σ (grey mesh). e, 2mFo−DFc (blue, contoured at 1.0 σ) and mFo−DFc (green/red, +/−3.5 σ) electron density maps of MT1-ramelteon (ligand purple, protein yellow) illustrating the small, unassigned electron density close to N2556.52 that is tentatively attributed to the essential additive 2-propan-ol. The distance from this electron density to the closest ligand atom is approximately 4.8 Å. f, Packing of MT1-PGS crystallised in the P 4 21 2 space group. The receptor is shown in green and the PGS fusion protein is shown in purple. g, Simulated annealing mFo−DFc omit maps (green mesh) of 2-pmt (orange sticks), 2-iodomelatonin (yellow), and agomelatine (cyan), contoured at 3.0 σ.