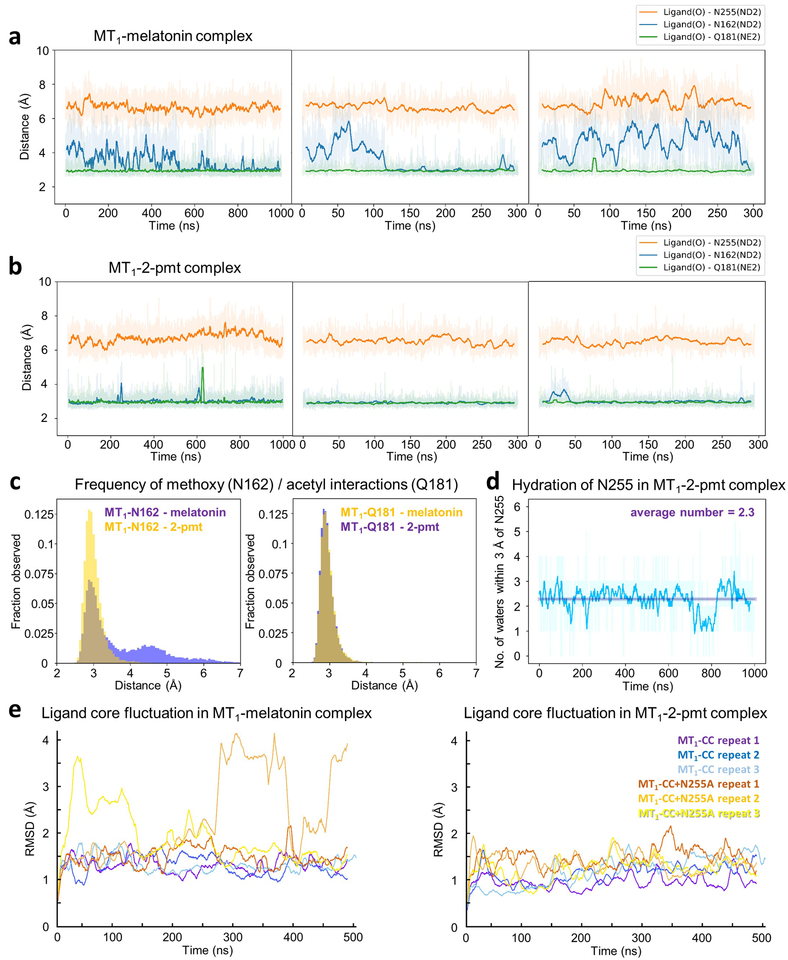
Extended Data Fig. 2 |. Molecular dynamics simulations.
a, b, Distance plots for interactions between residues in MT1 (N1624.60, atom type ND2 (Nδ); Q181ECL2, atom NE2 (Nε); N2556.52, atom ND2), and closest oxygen atoms of the methoxy and acetyl groups, respectively, in ligands melatonin (a) and 2-pmt (b) from three independent simulation runs. c, Distance histograms for interactions of methoxy with N162 (left), and Q181 with ligand acetyl tail (right), in melatonin and 2-pmt complexes. d, Hydration of residue N2556.52 over the course of a 1 μs simulation of the MT1-2-pmt complex from three independent simulations. e, Stability of ligand binding in simulations of MT1 complexes. Time dependence of RMSD for non-hydrogen atoms of melatonin shown for MT1-melatonin complex (left) and MT1-2-pmt complex (right). Three independent simulations of crystal construct (purple, blue, light blue) and crystal construct with N2556.52A mutation (orange, light orange, yellow) are shown, spanning 1.5 μs of cumulative time per system. Sampling rate was 10 frames per ns, and solid lines represent moving average values from 50 frames in all cases.