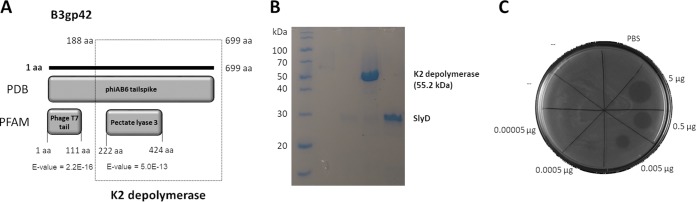
FIG 1.
K2 depolymerase identification. (A) In silico analysis of the A. baumannii phage vB_AbaP_B3 tail spike (B3gp42). The PBD database shows homology to the phiAB6 tail spike (5JS4_A), and PFAM identifies an N-terminal tail domain and a C-terminal depolymerase domain (pectate lyase 3, named K2 depolymerase) that was cloned (genetic region of bp 567 to 2100 of the B3_42 open reading frame). (B) SDS-PAGE gel demonstrating the expression and purification of hybrid protein expression (SlyD-K2 depolymerase) after TEV protease digestion to separate the K2 depolymerase from the SlyD fusion partner. (C) Drop tests of the K2 depolymerase onto NIPH 2061 strain.