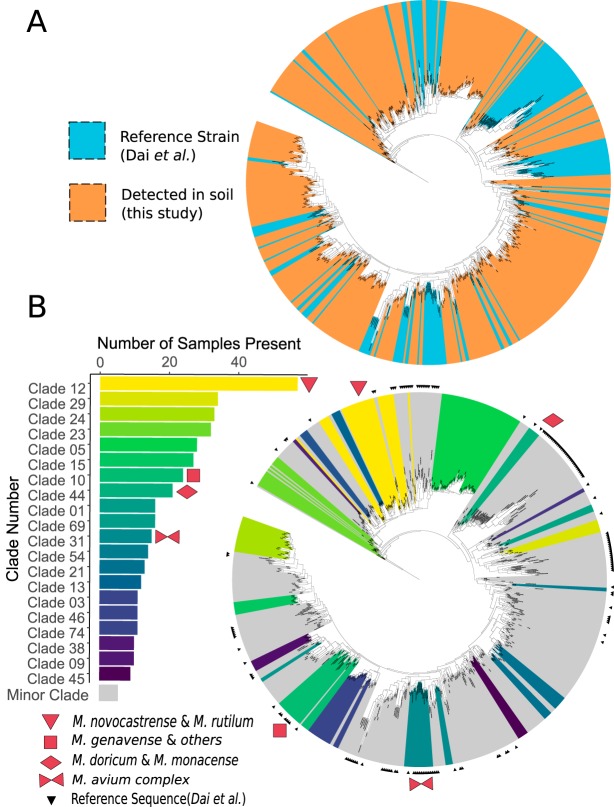
FIG 3.
Phylogenetic tree representing described and previously undescribed mycobacterial hsp65 sequences. (A) A total of 472 exact sequence variants (ESVs) were identified in soils sampled here, a majority of which represent novel and undescribed taxa. These ESVs span the known phylogenetic diversity of the genus. Colors indicate reference mycobacterial strains (blue) from Dai et al. (55) and sequences recovered from soils in this study (orange). (B) Closely related ESVs were grouped into 159 clades based on patristic distance. The top 20 most ubiquitous clades are highlighted in color, with yellow colors indicating higher ubiquity (clades found in more soil samples). Four of the clades included described members, as indicated by red symbols, namely, clade 10 (M. stomatepiae, M. genavense, M. florentinum, M. lentiflavum, M. montefiorense, and M. triplex), clade 12 (M. novocastrense and M. rutilum), clade 31 (including M. intracellulare, M. avium subsp. paratuberculosis, M. avium subsp. silvaticum, M. colombiense, M. chimaera, and M. avium subsp. avium), and clade 44 (M. doricum and M. monacense). Small black triangles mark sequences from the reference database. Both trees are rooted with the hsp65 sequence from Nocardia farcinica (DSM43665).