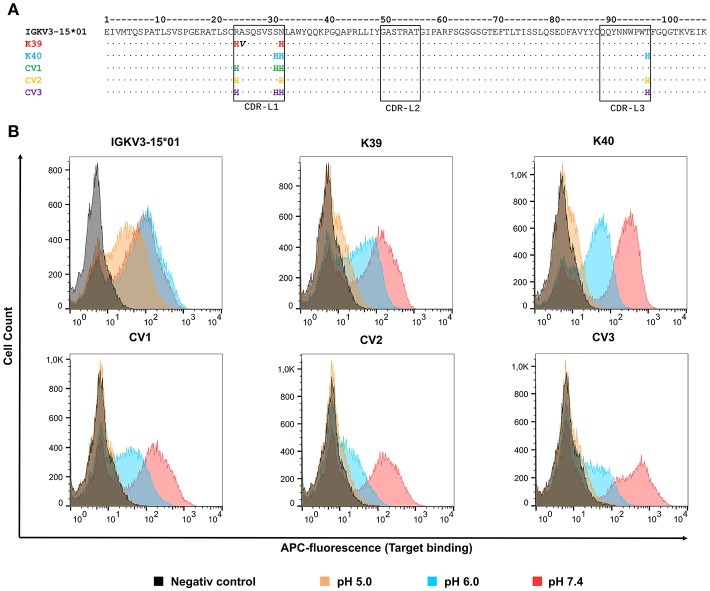
Figure 3.
Sequences and FACS analysis of the pH-responsive, His-doped common light chains. (A) Sequence alignment of the parental IGKV3-15*01 common light chains with the isolated pH-responsive light chains K39, and K40 as well as the permutated light chain variants CV1, CV2, and CV3. Histidines in CDR3 and CDR1 are highlighted in the respective light chain color. Non-histidine mutations are highlighted in bold black. The numbering refers to the Chothia numbering scheme. (B) Histograms of yeast cell single clone carrying the respective light chain in combination with the C5A heavy chain on the yeast surface. All cells, except the negative control which was incubated in PBS, were incubated with 62.5 nM CEACAM5 at their respective pH-value, and subsequently all cells were washed with PBSB at pH 7.4, and further labeled with anti-His and anti-mouse-APC. For each experiment 50,000 yeast cells were recorded.