Figure 2. MTX treatment depletes Bdnf mRNA and protein expression and disrupts TrkB signaling.
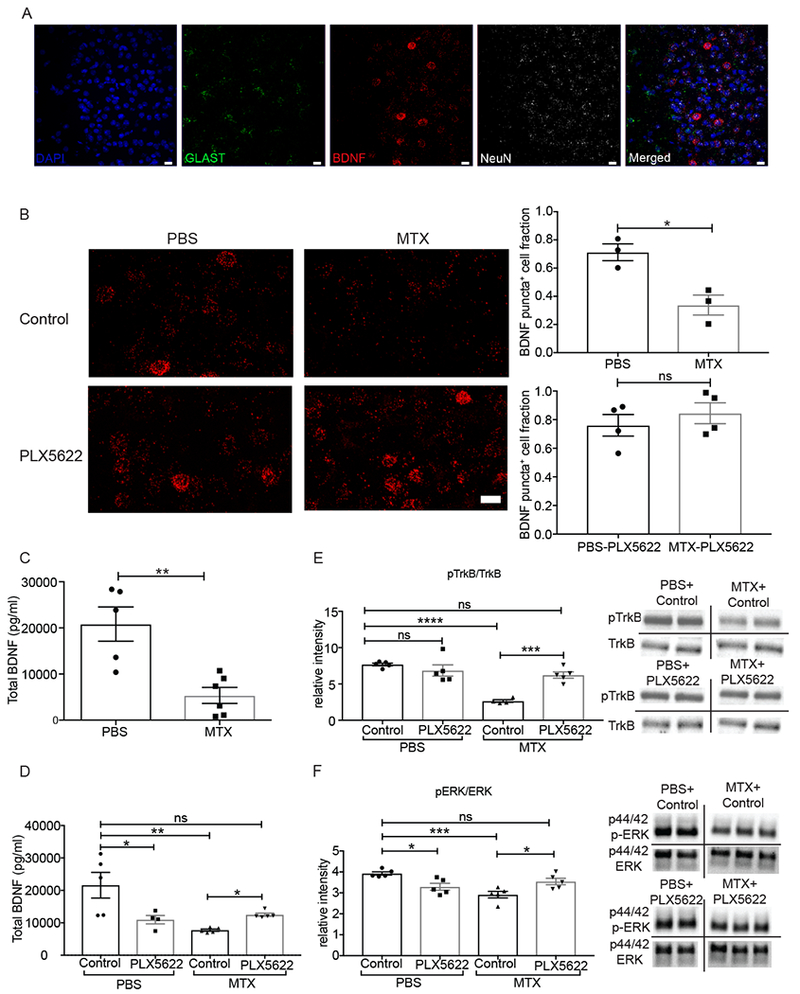
A) Representative image demonstrating RNAscope visualization of frontal cortex deep layer neurons (NeuN, white), astrocytes (Glast, green), and Bdnf mRNA (red). DAPI, blue. Scale bar = 20 μm
B) MTX treatment decreases Bdnf mRNA expression and microglial depletion with PLX5622 rescues Bdnf levels. (Left) Representative images of Bdnf puncta (red) in mice treated with PBS+control chow (n=3 mice) and PBS+PLX5622 chow (n=4 mice), MTX+control chow (n=3 mice) and MTX+PLX5622 chow (n=4 mice). (right) Quantification of DAPI+ cells expressing above threshold Bdnf mRNA puncta in each group. Scale bar = 20 μm
C) Decreased Bdnf protein levels following MTX exposure. Total Bdnf protein levels from frontal deep cortex and corpus callosum tissue microdissected at P63 and measured by ELISA in mice treated with MTX (n = 6 mice) or PBS vehicle control (n = 5 mice).
D) Microglial depletion with PLX5622 restores Bdnf levels following MTX exposure. Total Bdnf protein levels from frontal deep cortex and corpus callosum tissue microdissected at P63 and measured by ELISA in mice treated with PBS+control chow (n = 5 mice), PBS+ PLX5622 chow (n = 4 mice), and MTX+control chow (n = 5 mice), MTX+PLX5622 chow (n = 5 mice).
E) Microglial depletion with PLX5622 restores TrkB signaling following MTX exposure. Ratio of phospho-TrkB/TrkB western blot band intensity from microdissected frontal deep cortex and corpus callosum tissue of mice treated with PBS+control chow (n = 5 mice), PBS+ PLX5622 chow (n = 5 mice), MTX+control chow (n = 4 mice), and MTX+PLX5622 chow (n = 5 mice). Representative images of westerns to the right of the graph.
F) Microglial depletion with PLX5622 restores downstream TrkB signaling following MTX exposure. Ratio of phospho-ERK/ERK western blot band intensity from microdissected frontal deep cortex and corpus callosum tissue of mice treated with PBS+control chow (n = 5 mice), PBS+ PLX5622 chow (n = 5 mice), MTX+control chow (n = 5 mice), and MTX+PLX5622 chow (n = 5 mice). Representative images of westerns to the right of the graph.
Data shown as mean ± SEM. Each point = one mouse. ns = p > 0.05, * p < 0.05, ** p < 0.01, *** p<0.001, **** p<0.0001. Students t-tests (B, C); Two-way ANOVA with Tukey post-hoc analysis for multiple comparisons (D,E,F).
