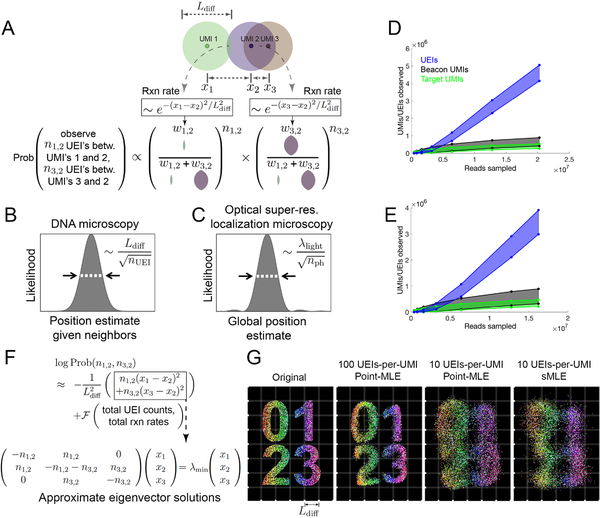
Figure 3. Image inference from DNA microscopy data.
(A) Modeling diffusion of amplifying UMIs as isotropic across length scale Ldiff allows the likelihood of a UMI-position solution to be evaluated given observed UEI counts. (B,C) Uncertainty in DNA vs. super-resolution microscopy. Given its reacting partners’ positions, DNA microscopy (left) defines a UMI’s uncertainty as a physical length scale (DNA diffusion distance, Ldiff) divided by the square-root of the number of individual quanta measured (UEIs) in a manner analogous to quanta (photons) in super-resolution microscopy (right). (D,E) Rarefaction of UMI and UEI data. Shown are curves with an upper-bound, indicating total UMI/UEI counts, and a lower-bound, indicating those from the final pruned UEI matrix, for samples 1 (D) and 2 (E). (F) The sMLE algorithm uses eigenvector solutions to part of the position-probability function to identify a linear basis for the solution to the full likelihood function. (G) sMLE enhances performance in free-diffusion simulation tests. From left: original image, results from point-MLE on simulated images with 100 or 10 UEIs/UMI, and from sMLE with 10 UEIs/UMI.