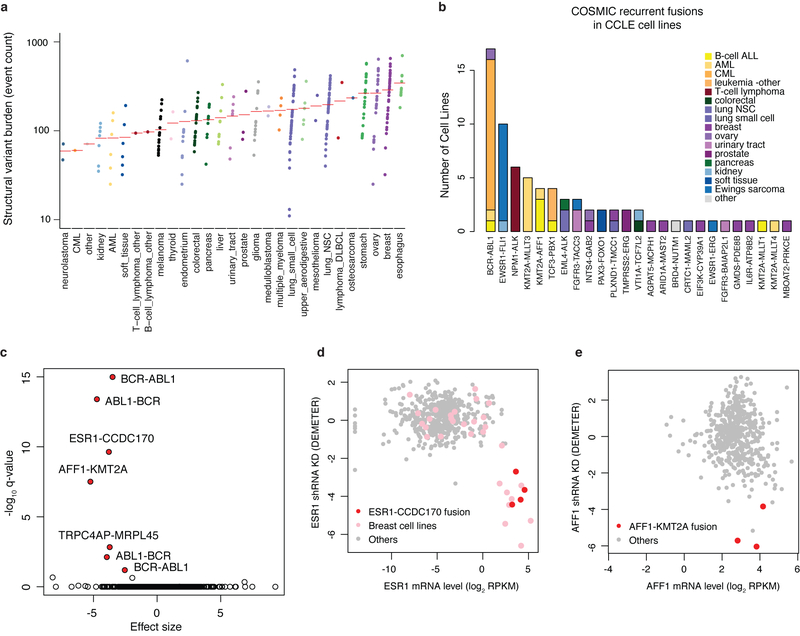
Extended Data Fig. 3. Annotation of structural variants and fusions in CCLE cell lines.
a, Structural variant (SV) burden in CCLE whole genomes. Structural variants detected by SvABA in cell lines grouped by tissue type are plotted in the order of mean structural variant burden (red bar in each facet). b, Bar plot of recurrent COSMIC fusions detected in CCLE RNA-seq data coloured by cell line lineage. c, Volcano plot of Achilles RNAi gene dependencies versus CCLE fusions for cell lines (n = 478) common between CCLE and Achilles datasets. P values determined by two-sided t-test. Genes with significant adjusted P values (false discovery rate (FDR) < 0.1) are highlighted. d, e, Examples of fusions associated with gene dependency: cell lines with ESR1-CCDC170 fusion (n = 4) are sensitive to ESR1 shRNA knockdown (d), and cell lines with AFF1-KMT2A fusion (n = 3) are sensitive to AFF1 shRNA knockdown (e). The x axis shows mRNA expression, and the y axis shows Achilles RNAi gene dependency DEMETER score5.