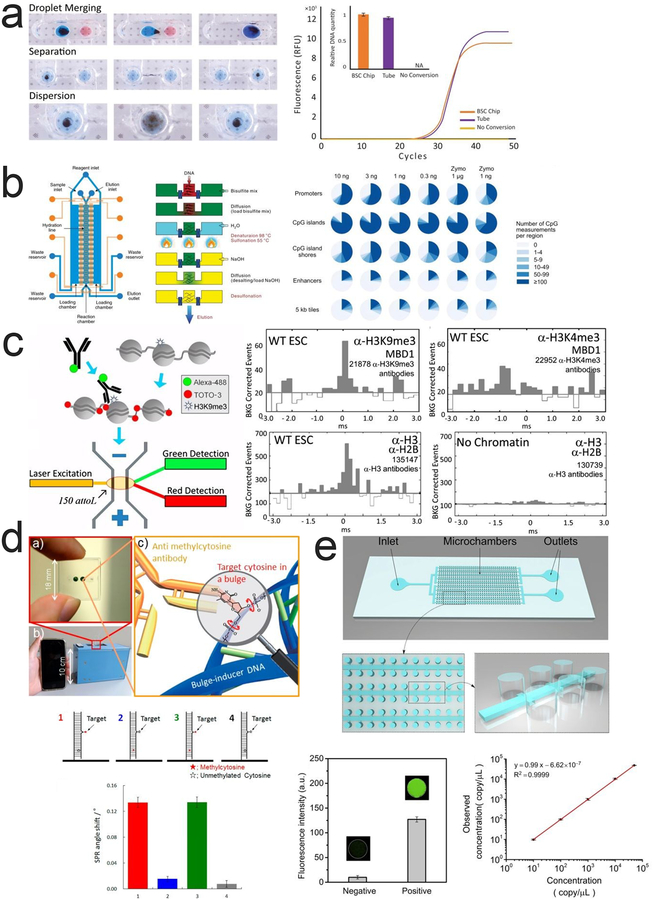
Figure 3 |. Microfluidic technologies for profiling DNA methylation.
a. Steps required for on-chip bisulfite conversion. Droplet merging, separation and mixing are performed through magnetic actuation of magnetic beads. Reprinted with permission159. Copyright 2016 Springer Science+Business Media New York. b. MID-RRBS microfluidic device with a fluidic layer (blue) and a control layer (orange) and a cross-sectional view of the reaction and loading chambers. Pie chart shows MID-RRBS and Zymo commercial kit coverage of gene promoters, CpG islands, CpG island shores, enhancers and 5kb tiles. Reprinted with permission118. Copyright 2018 Macmillan Publishers Ltd. c. SCAN workflow and simultaneous detection of DNA methylation and histone modifications. Reprinted with permission134. Copyright 2013 National Academy of Sciences. d. Immunoreaction between antibody and target cytosine causing SPR angle change. Reprinted with permission161. Copyright 2015 American Chemical Society. e. The microfluidic device for using digital PCR to detect PCDHGB6 for examining methylated DNA with different concentrations. Reprinted with permission162. Copyright 2017 Elsevier.