SUMMARY
To what extent do structural and biophysical features of food allergen proteins distinguish them from other proteins in our diet? Invertebrate tropomyosins (Tpm) as a class are considered ‘pan-allergens’, inducing food allergy to shellfish and respiratory allergy to dust mites. Vertebrate Tpms are not known to elicit allergy or cross-reactivity, despite their high structural similarity and sequence identity to invertebrate homologs. We expect allergens are sufficiently stable against gastro-intestinal proteases to survive for immune sensitization in the intestines, and that proteolytic stability will correlate with thermodynamic stability. Thermal denaturation of shrimp Tpm shows that it is more stable than non-allergen vertebrate Tpm. Shrimp Tpm is also more resistant to digestion. Molecular dynamics uncover local dynamics that select epitopes, and global differences in flexibility between shrimp and pig Tpm that discriminate allergens from non-allergens. Molecular determinants of allergenicity depend not only on sequence but on contributions of protein structure and dynamics.
eTOC Blurb
Predictive models that anticipate new food allergens based on sequence homology to existing allergens, do not perform well on the shellfish allergen tropomyosin, which shares high identity with non-allergenic vertebrate forms. Instead, allergens are discriminated from non-allergens by biophysical properties and structural dynamics.
Graphical Abstract
INTRODUCTION
After each meal, immune cells lining the gastrointestinal tract interact with countless food proteins with diverse sequences and structures. The normal function of these immune cells is to distinguish food from enterotoxins and pathogens. However, failure to do so results in misidentification of benign food proteins as antigens leading to sensitization for a later allergenic response. Despite the variety of proteins consumed, just a handful are responsible for food allergies. 2% of domain families as defined by Pfam (Finn et al., 2014) are associated with IgE mediated allergies in genera. Of these, approximately ten families account for about half of all documented allergenic proteins (Tyagi et al., 2015). Food allergies display diverse clinical manifestations indicating a complex multi-faceted mechanism for pathogenesis. The gastro-intestinal mucosal barrier plays an important role in this process by preventing the entry of harmful entero-pathogens (Sampson, 1999). During the course of postnatal development, food allergies can result from a lack of maturation in the intestinal mucosal barrier, or a dysregulation of the immune cells to bias responses toward hypersensitivity (Sicherer and Sampson, 2010).
Multiple studies have tried to relate protein sequence to allergenicity. Sequence similarity of a protein to the metazoan parasite proteome, which IgE mediated immunity is evolved to protect against, may provide one factor (Fitzsimmons and Dunne, 2009). Evolutionary distance from human homologs may also correlate with protein allergenicity (Jenkins et al., 2007). These phylogenetic studies identify a sequence identity cutoff between allergenic and non-allergenic proteins within the same family to be approximately 60%. This is well above the 35% identity threshold established by the FAO/WHO guidelines for genetically modified foods as a precaution against allergic cross-reactivity (Taylor, 2002). The fact that relatively similar sequences may diverge in their allergenicity indicates other factors may be involved. In the present study, we aim to better understand this divergence in the context of protein structure.
Current bioinformatics tools used to assess the allergenicity of a protein rely primarily on protein sequence analysis, assessing whether a novel protein will cross-react with IgEs developed against known allergens (Ladics et al., 2011; Taylor, 1997). FAO/WHO guidelines acknowledge the limitations of these methods for identifying high-risk proteins that may cause food allergy and recommend additional measures including serum testing against individuals with reactions against similar allergens, as well as assays to demonstrate resistance to pepsin digestion (Commission et al., 2003).
Proteins known to cause food allergies are often more resilient to gastrointestinal proteases than non-allergenic ones, indicating a difference in stability that enables a greater likelihood of sensitization (Astwood et al.). For example, mouse models show the use of antacids increase sensitization to food allergens, potentially by neutralizing gastric proteases (Pali-Scholl and Jensen-Jarolim, 2011; Untersmayr et al., 2003). The peanut allergen, Ara h 2, contains inhibitory sequences to trypsin, resulting in a decrease in its digestibility (Maleki et al., 2003). However, other studies have challenged this correlation between protein digestibility and protein allergenicity (Fu et al., 2002; Herman et al., 2007). A structural comparison between allergenic and non-allergenic proteins may provide insight into the differences that impact digestion.
We use Tropomyosin (Tpm) as a model for understanding how allergenicity relates to structure and gastro-intestinal stability. As the primary allergen in shellfish, it causes one of the most common food allergies affecting 2% of the US adult population (Sicherer et al., 2004). While invertebrate Tpm is well associated with food allergy, few clinical examples exist of vertebrate Tpm associated allergies (Ayuso et al., 1999; Gonzalez-Fernandez et al., 2016; Reese et al., 1999). Five major IgE epitopes were subsequently mapped in the related Penaeus aztecus Tpm using a peptide-tiling array assay testing IgE binding from patient sera and defined by the following residue ranges: epitope 1 (residues 43 – 57), epitope 2 (residues 85 – 105), epitope 3 (residues 133 – 153), epitope 4 (residues 187 – 201), epitope 5 (residues 247 – 284) (Ayuso et al., 2002; Reese et al., 2001; Reese et al., 2005). IgE binding regions ranged from 14 to 36 residues in length and were separated by approximately 40–50 residues, spanning the entire Tpm molecule. P. aztecus epitopes cross-reacted to other known invertebrate allergens such as Tpms from lobster, cockroach or dust mites (Ayuso et al., 2002; DeWitt et al., 2004). However, protein sequence does not clearly correlate with allergenicity. Tpm sequences are strongly conserved and current tools that use sequence based algorithms for predicting allergenicity cannot discriminate between allergenic and non-allergenic species (Ladics et al., 2011). When comparing distributions of sequence similarities between epitope and non-epitope regions within allergenic sequences, no significant differences are found as well (Reese et al., 2001).
The linear topology of Tpm facilitates modeling of sequence, structure and dynamics. As a continuous coiled-coil parallel homodimer, the structure of Tpm allows for only local contact within a sequence as it follows a repetitive ‘abcdef’ heptad pattern (Lupas and Gruber, 2005; McLachlan et al., 1975; Whitby and Phillips, 2000) (Fig. 1A). Classically within a coiled-coil, positions are solvent exposed except the a and d residues, which form a hydrophobic oligomer interface. Sequence and structural deviations in Tpm from an idealized coiled-coil impart inhomogeneity and flexibility necessary for proper function of the protein (Brown et al., 2001; Hitchcock-DeGregori, 2008; Singh and Hitchcock-DeGregori, 2006). This results in a complex folding pathway with multiple transitions seen in thermal denaturation (Ishii et al., 1992; Singh and Hitchcock-DeGregori, 2003). Subtle sequence changes such as point mutations, for example G126A and E180G in rat Tpm, have dramatic effects on stability and proteolytic susceptibility (Ly and Lehrer, 2012; Matyushenko et al., 2015; Nevzorov et al., 2011). This motivates us to explore differences between allergenic and non-allergenic forms of Tpm at the structural level in addition to sequence.
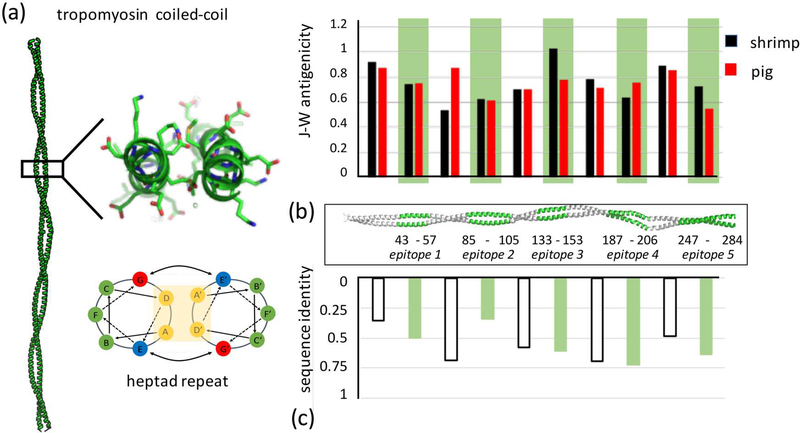
Figure 1:
(a) Tropomyosin forms an extended parallel coiled-coil dimer that consists of approximately 40 uninterrupted heptad repeats, a regular pattern of seven residues whose position can be described as a to g based on their position along the helix. Structure shown is pig Tpm - PDB ID 1C1G (Whitby and Phillips, 2000). (b) The five major epitopes identified in previous studies (Ayuso et al., 2002) are highlighted in green along the coiled coil. The J-W index computes antigenicity based on sequence. For shrimp Tpm, there is no statistical difference between epitope and non-epitope antigenicities (student’s t-test p = 0.86), or between shrimp and pig epitope regions (p = 0.50). (c) Cross-reactivity has not been observed between shrimp and pig Tpms. This is likely not due to lower sequence identity within epitopes versus non-epitope regions (p = 0.99).
For the current study, pig and shrimp homologs are used as exemplars of non-allergenic and allergenic Tpm respectively. They share 56% sequence identity and 70% similarity with no gaps (Fig. 1B, C). Traditional methods of measuring antigenicity, such as the Jameson-Wolf index, compiles structural features from sequence such as surface probability, hydrophobicity, backbone flexibility and secondary structure prediction, as an indicator of antigenic potential (Jameson and Wolf, 1988). Comparing both sequence homology and antigenicity in pig and shrimp Tpm reveals no clear correlations with known epitopes. This suggests that a more detailed structural understanding is needed to understand differences in stability and digestibility between allergenic and non-allergenic homologs. Using a combination of proteomic, biophysical and molecular simulation approaches we identify consistent differences in structural stability and dynamics between allergenic epitopes and non-epitopes in Tpm.
RESULTS AND DISCUSSION
Simulating gastric and intestinal digestion
Laboratory digestion simulations were performed on purified Tpms from natural sources. Penaeus aztecus Tpm was isolated and purified from frozen shrimp purchased at a local supermarket. MS/MS analysis of peptide fragments from pepsinolysis confirmed full coverage Tpm corresponding to the P. aztecus sequence. Sus scrofa (porcine) Tpm was purchased from Sigma Chemicals (St Louis, MO) and further purified to remove lower molecular weight contaminants. Both proteins were subject to simulated gastric digestion adhering to standardized protocols for assessing food safety (Commission et al., 2003) as described in Methods. Briefly, Tpm was incubated in pepsin and HCl at pH 2.0, 37°C to mimic the gastric environment. Aliquots were extracted at a series of time points for analysis and pepsin inactivated by raising the pH. For simulated intestinal digestion, one-hour and two-hour gastric digestion products were adjusted to pH 7.5–7.9 and allowed to digest in a solution of chymotrypsin and trypsin, subsequently quenching with the serine protease inhibitor Pefabloc SC.
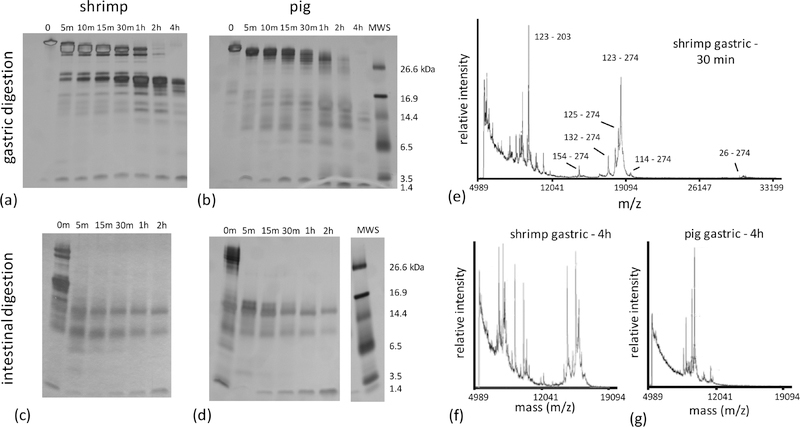
Under simulated gastric conditions, shrimp Tpm demonstrated resistance to digestion, with proteolysis proceeding through a series of intermediate molecular weight (MW) species (Figure 2a). MALDI-TOF analysis of digestion products showed rapid cleavage of the N- and C-termini of full-length shrimp Tpm, producing a major product consisting of residues 26–274. A subsequent cleavage in the middle produced a population of ~18 kDa shrimp Tpm C-terminal fragments. The primary product had a mass corresponding to residues 123–274, with others starting at nearby positions and ending at 274 (Figure 2e), which persisted throughout the extended four-hour experiment. Notably, similarly sized N-terminal fragments were not identified in analysis of the MALDI-TOF mass spectra suggesting preferential survival of the C-terminal half of shrimp Tpm. A sequence of proteolytic events occurs starting with immediate cleavage of residues 1–25 and 275–284 and a subsequent cleavage at position 123 prior to further fragmentation.
Figure 2:
Time series of digestion under gastric conditions were performed for Tpm purified from (a) shrimp and (b) pig muscle tissue. Digestion products were separated with SDS page and visualized with silver staining. Similarly, products after 1 hour of simulated gastric digestion of (c) shrimp and (d) pig Tpm were further fragmented under enzymatic conditions corresponding to the intestinal phase. Gels have been cropped to eliminate superfluous lanes – cropping sites are indicated by white space between lanes. (e-g) Residues corresponding to dominant fragments surviving gastric digestion were identified with from MALDI-TOF spectra. The dominant fragment in shrimp is approximately 18 kDa, and consists of residues 123 to 274.
Pig Tpm also exhibited significant resistance to proteolysis with major high MW species evident through one hour of simulated gastric digestion (Figure 2b). Although some intermediate MW species are evident in the digestion, peaks in the MALDI-TOF mass spectra could not be uniquely attributed to specific species. Unlike shrimp Tpm, at four hours, little of these intermediate MW species in the pig homolog are detected either by SDS-PAGE or mass spectrometric analysis (Figure 2f,g).
Despite high sequence similarity, the patterns of gastric digestion are different for the two forms of Tpm. Shrimp Tpm shows significantly greater resistance to simulated gastric digestion than pig Tpm. Furthermore, shrimp Tpm digestion proceeds through a persistent C-terminal half intermediate, whereas similarly long-lived species are not clearly observed for pig Tpm.
In ideal cases, non-allergens are rapidly digested in seconds to minutes (Astwood et al., 1996). The differences between shrimp and pig Tpm digestion are not as clear. Intermediate MW species persist in both digestions after two hours, the upper limit for standard gastric residence times (Camilleri et al., 1989). Simulated intestinal digestion profiles are also similar (Figure 2c,d) in both shrimp and pig experiments. Further examination of differences in the digestion, stability and structure of the two Tpms was warranted to gain further insight into their allergenic potential.
To examine whether digestion selects for small MW fragments of Tpm corresponding to observed epitopes, we used a high-resolution mass spectrometry approach. Gastric and intestinal digestion was simulated in vitro as above. Peptide fragments were identified by LC-MS/MS, where the spectral count was used to quantify the relative abundance of a peptide over the time series. Spectral counts are not necessarily reliable for quantitative comparison of abundances between peptides of different chemical compositions (Lundgren et al., 2010). Therefore, we have focused our analysis to normalization of relative abundance independently for each peptide sequence.
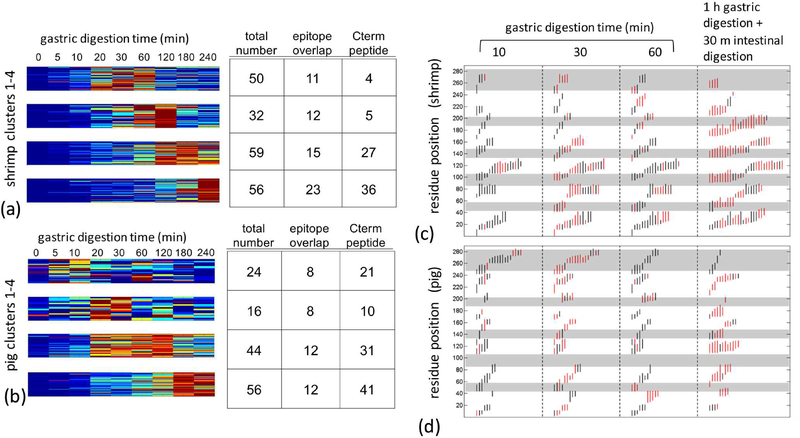
In shrimp Tpm, approximately two-hundred fragments were identified during gastric digestion, corresponding to full coverage of the protein sequence. Peptides identified later in digestion were inferred to be in regions resilient to enzymatic cleavage. To show this, fragment abundances were clustered into four groups based upon the time of peak relative abundance. Peptides from the C-terminal half of the protein are more represented in the cluster of long surviving peptides than the N-terminal half. In addition, the number of peptides overlapping the major epitopes increases in groups with fragment survival (Figure 3a). Pig Tpm overall produced fewer fragments identifiable by MS/MS. Similar to shrimp, fragments that originated in the C terminal half of the protein were more concentrated in long surviving clusters. Unlike shrimp, no correlation was observed between persistence and epitope overlap (Figure 3b).
Figure 3:
Fragments derived from a time course of shrimp and pig Tpm simulated gastric digestion was quantified by LC-MS/MS. Approximately 200 fragments were identified across the digestion time from 5 min to 4hrs. (a) Shrimp and (b) pig Tpm fragments were quantified based on relative abundance across the time series, where abundance is represented by a rainbow palette from blue-low to red-high. Peptides were grouped by k-means clustering. For each cluster, the total number of peptide, and number of peptides overlapping the 5 epitope regions and C terminal half of the protein defined as residues 123–284 are tabulated on the right. Sequence position and relative are shown for (c) shrimp and (d) pig Tpm. Maximum relative abundance of a specific fragment during gastric and intestinal digestion time-course are indicated in red. Grey bands demarcate epitopes.
When simulated intestinal digestion phase was added in tandem to simulated gastric digestion, shrimp Tpm was found to have twice the number of surviving peptides generated and five-fold greater overlap with epitope regions than the pig homolog (Figure 3c,d). These observations from high resolution MS/MS indicate that shrimp Tpm allows for greater survival of fragments from proteolysis particularly at areas that overlap with common IgE epitopes.
Distinct patterns of proteolytic susceptibility were observed between the two Tpms as well as between epitope and non-epitope regions. These patterns likely arise from the relationship between thermodynamic stability and proteolytic susceptibility. This was explored by probing differences in global and local stability in pig and shrimp forms of Tpm.
Understanding digestion in terms of stability
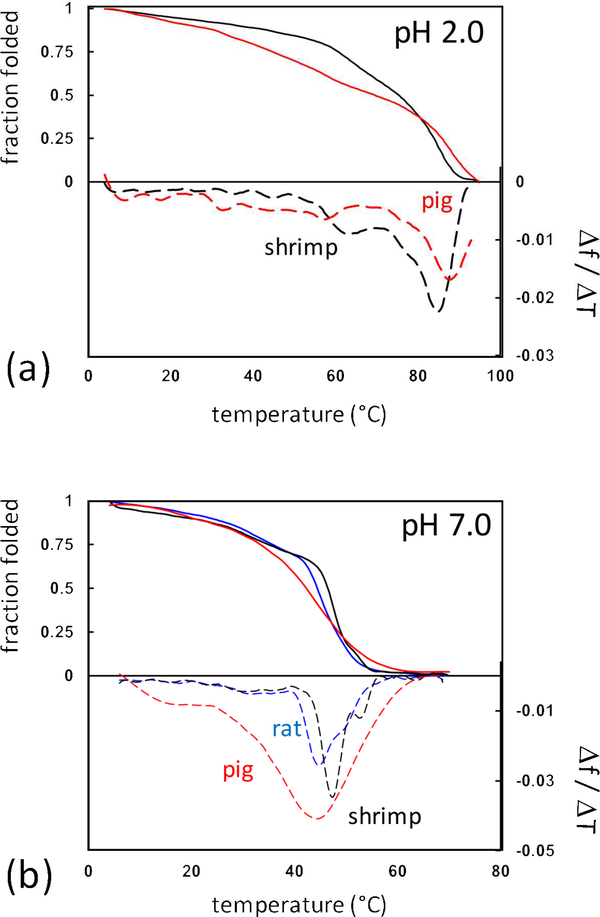
The rate of digestion was expected to be related to local stability and structure as regions that are well structured are generally protected against enzymatic cleavage (Park and Marqusee, 2004). CD spectroscopy was used to measure loss in α-helicity upon thermal denaturation (Figure 4a). Under acid pH, both shrimp and pig Tpm revealed a remarkably high dominant transition at 85°C and 88°C respectively. Similar stabilities have been observed with other vertebrate Tpms under acidic conditions (Lehrer and Yuan, 1998; Williams and Swenson, 1981; Woods, 1977). Proteins tend to be most stable near their isoelectric point, and Tpms in general have isoelectric points less than 5 (Lehrer and Yuan, 1998; Reese et al., 1999; Talley and Alexov, 2010). The high stability of both Tpms at acidic pH may explain their resistance to in vitro gastric digestion. This is in contrast with the behavior of the peanut allergen Ara h 1, which adopts a molten-globule state that is rapidly degraded under gastric conditions (Khan et al., 2013; Ohgushi and Wada, 1983).
Figure 4:
Thermal denaturation of Tpm was observed by CD spectroscopy following helical transitions at 222nm. Thermal denaturation (solid lines) and first-derivative plots were calculated (dashed lines) of pig (red), shrimp (black), and rat (blue) Tpm at (a) acidic and (b) neutral conditions. Temperature ranged from 4°C to 70°C or 95°C for neutral and acidic conditions respectively.
Based on this in vitro model, significant digestion appears to occur in the intestinal phase under neutral pH conditions. Previous work examining Tpm unfolding has shown multiple transitions under neutral conditions (Ishii et al., 1992; Potekhin and Privalov, 1982). Consistent with this, multiple transitions were seen for both pig and shrimp upon thermal denaturation monitored by CD (Figure 4b). The unusually broad transitions in pig Tpm may be due to the fact that it was extracted from skeletal muscle tissue containing multiple isoforms, resulting in a heterogeneous population of coiled-coil dimers (Geeves et al., 2015). Alternatively, a single isoform vertebrate preparation can be produced using the well-studied recombinant rat Tpm construct. Comparing rat and shrimp we find the characteristic three folding transitions. The two dominant transitions in shrimp Tpm were at higher temperatures than the rat construct. Changes in the positions of folding transitions have been demonstrated to alter the pattern of proteolysis in previously studied vertebrate point mutations (Ly and Lehrer, 2012; Nevzorov et al., 2011; Sumida et al., 2008). The increased stability of shrimp Tpm at neutral pH may account for the differences we observed in intestinal digestion.
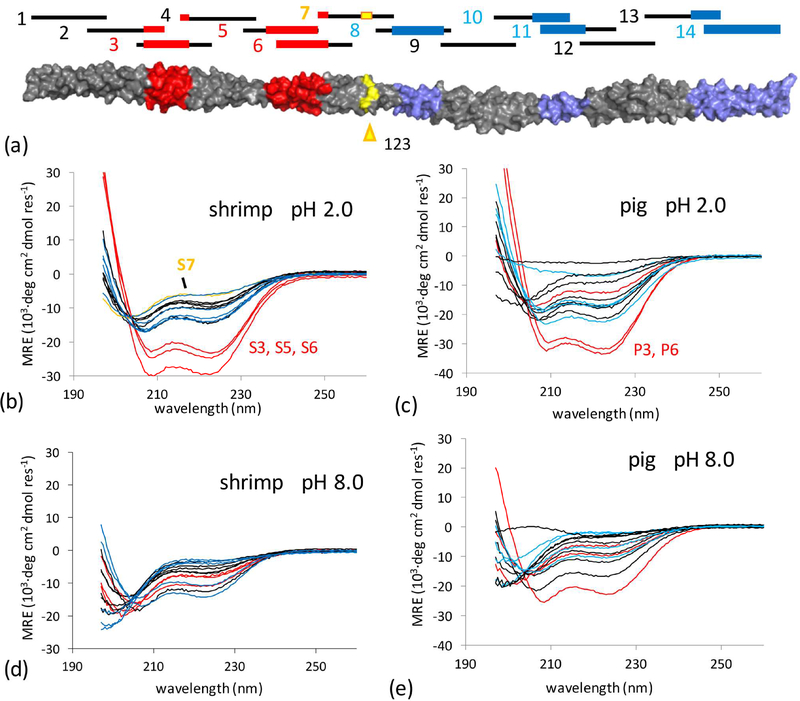
We expect the epitopes of shrimp Tpm to correspond to high local stability. The resilient C-terminal half of shrimp Tpm to gastric digestion has already suggested a dichotomy in conformational stability between the N and C terminal regions that separates homologs. In order to further explore local stability, we constructed fourteen peptides that spanned the Tpm coiled-coil (Figure 5a). One concern is that peptide fragments in isolation may adopt other structures beside an α-helical dimeric parallel coiled coil (i.e. antiparallel association of helices, or trimer, tetramer, etc.) (Harbury et al., 1993). To constrain conformation, a nine-residue leader sequence from the yeast transcription factor GCN4 was added to each peptide, where Asn 8 in the sequence forms an intermolecular hydrogen bond that specifies a parallel dimer (Figure S1) (O’Shea et al., 1991; Zeng et al., 1997). This strategy has been used multiple times in the study of Tpm, where presence of the intermolecular Asn-Asn hydrogen bond has been confirmed in high-resolution structures (Greenfield et al., 2001; Li et al., 2002; Murakami et al., 2008).
Figure 5:
(a) 14 peptides were synthesized spanning Tpm sequence for either shrimp or pig proteins. Epitopes in red are prior to the initial pepsin cleavage site at residue 123 and defined as part of the N terminal half (NTH) of Tpm, conversely, epitopes downstream of the cleavage site are in the digestion-resistant (see Fig 2) C terminal half (CTH). (b – e) CD spectra of designed peptides from pig and shrimp Tpm sequences under neutral and acidic conditions. Spectra of peptides with overlap to epitopes within the NTH and CTH were colored red and blue respectively. Residue 123 in shrimp Tpm is suggested to be near the initial cleavage site for shrimp Tpm, peptide S7 overlaps it and is highlighted in yellow.
CD spectra were obtained under gastric conditions (pH 2) to measure local structure. Only peptides S3, S5, and S6 in shrimp had significant helical structure (Figure 5b). These peptides correspond to the first two epitopes in the N terminal half of the protein. Corresponding peptides P3 and P6 from pig were also helical (Figure 5c). The remaining peptides across both pig and shrimp Tpm had little to no helical structure. Notably peptide S7 which contains the initial cleavage site at position 123 is the least structured in shrimp suggesting its rapid proteolysis from local structural instability. In contrast in pig Tpm, peptide P7 maintained significant helical structure. Under neutral pH, all peptides from shrimp and pig were observed to be less structured (Figure 5d,e), which corresponds to the dramatic decrease in global stability between acidic and neutral conditions for full length Tpm. In shrimp Tpm, all peptides were unstructured at neutral pH.
Together, these observations suggest a complex model relating epitope stability and digestibility. In the N-terminal domain, synthetic peptides spanning epitopes demonstrated significant local structure that may explain their persistence in digestion. On the other hand, peptides corresponding to the three C terminal epitopes lack local structure. Their persistence instead may be due to the proteolytic survival of the larger C terminal region as a whole.
Contributions of structure and dynamics
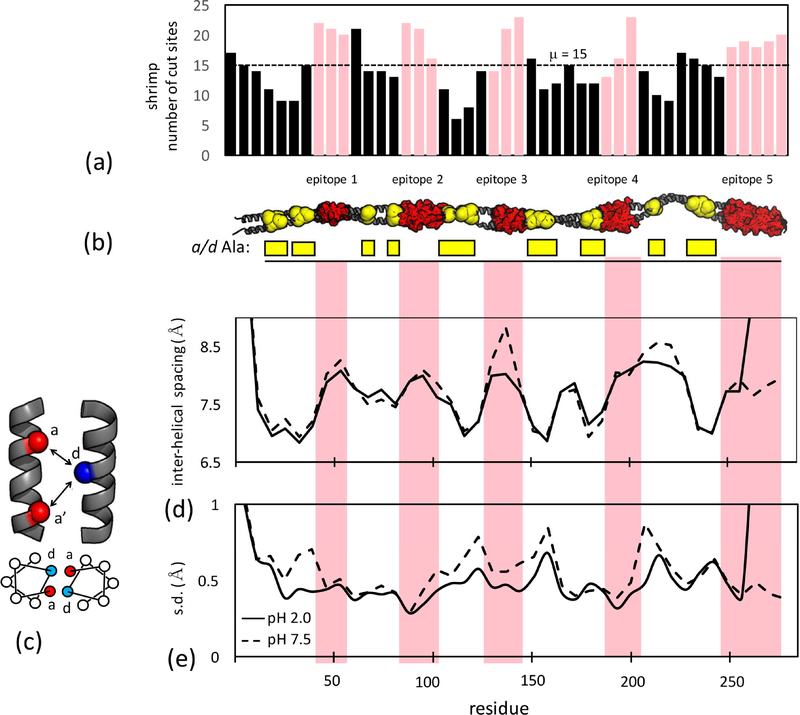
An apparent contradiction exists between observed and predicted proteolytic susceptibility of Tpm. One would expect the number of predicted cleavage sites based on known specificities of pepsin, trypsin and chymotrypsin to be lower within epitopes. Instead, the opposite is found (Figs. 6A, S2). Epitopes in both shrimp and pig Tpm consistently contain a higher-than-average number of cleavage sites relative to the intervening regions. This is due to the existence of periodic Ala-clusters, groups of alanine at core ‘a’ and ‘d’ positions in the helix heptad (Singh and Hitchcock-DeGregori, 2003, 2006). Pepsin, trypsin and chymotrypsin prefer larger hydrophobic residues such as Phe or Leu at the P1 position N-terminal to the scissile bond. Gastrointestinal digestion patterns of Tpm cannot be explained based on sequence-derived proteolysis susceptibility.
Figure 6:
(a) Number of pepsin, trypsin and chymotrypsin cleavage sites for 16-aa fragments of shrimp Tpm starting every six residues (i.e. 1–16, 7–22 …). Pink bars indicate fragments that overlap epitopes. (b) Epitopes (red) and a or d heptad position Ala-clusters (yellow) highlighted on a model of the Tpm coiled coil. (c) interhelical spacing is calculated between the d position on one chain and the a and a’ postions on the neighboring chain as (da + da’)/2. (d) mean interhelical spacing over the shrimp Tpm molecular dyanmics trajectory at acid and neutral pH. (e) standard deviation of interhelical spacing indicating local flexibility of core packing. See also Table S1, Figures S2, S3, S4, S6.
Periodic Ala-clusters are conserved across vertebrate and invertebrate Tpms (Barua et al., 2011). The small, nonpolar core alanines are proposed to allow local helical flexibility that facilitates binding to actin, and bending of Tpm as it wraps around the actin filament (Brown et al., 2005; Zheng et al., 2016). Molecular simulations of vertebrate Tpms show closer helix-helix packing and larger root-mean-square fluctuations at Ala-clusters. If these properties extend to invertebrate Tpms under both low and neutral pH conditions, local variations in dynamics may likely best describe proteolytic susceptibility. Proteolytic susceptibility is related to structural dynamics; local transient unfolding would likely allow for a higher probability of cleavage (Park and Marqusee, 2004). Modeling Ala-clusters and epitopes along the Tpm coiled-coil clearly shows these two features arrayed in mutually-exclusive, alternating domains (Fig. 6B).
We simulated shrimp and pig Tpm choosing protonation states for titratable groups that reflected either acidic (pH 2.0) or neutral (pH 8.0) conditions. Under acidic conditions both Tpms are more helical than neutral (assessed by occupancy of i, i+4 hydrogen bonds), consistent with experimental stabilities (Table S1, Fig. S3). Ala-clusters facilitate close interhelical spacing of core residues (Fig. 6C, D) for shrimp Tpm under both acid and neutral conditions. One significant deviation is noted in shrimp Tpm in the center of epitope 3 where the interhelical spacing expands at neutral pH. This is potentially due the presence of Asp 134 in the helix-helix interface, driving expansion due to charge-charge repulsion in the unprotonated state. These structural features were largely similar for pig Tpm (Fig S4).
Ala-cluster regions in shrimp Tpm coincided with regions of elevated flexibility (Fig. 6E, S4), while epitopes showed less variation in interhelical spacing. Flexibility is largely anti-correlated with interhelical spacing, indicating that packing of larger hydrophobic residues stabilizes the local fold under both gastric and intestinal conditions. The increased flexibility of Ala-clusters is a plausible mechanism of their preferential proteolysis, promoting digestive survival of epitope containing fragments.
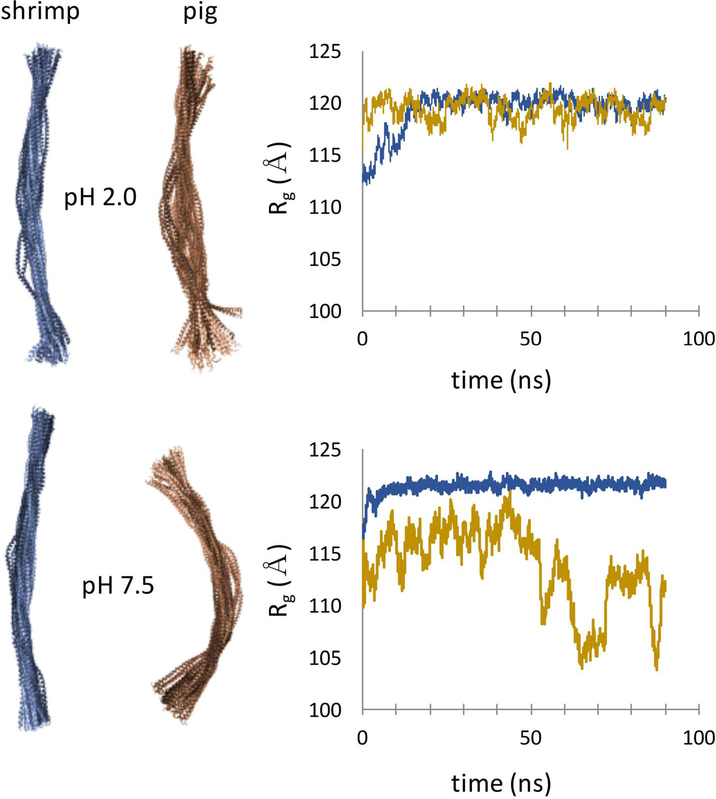
The heterogeneity of local dynamics provides little discrimination between allergen and non-allergen forms of Tpm. Ala-cluster effects on dynamics are seen in both shrimp and pig simulations, consistent with the similar stabilities of peptides S1–14 compared to P1-P14. The largest differences are seen in order parameters that describe global structural features, such as radius of gyration, Rg (Fig. 7, S5). In the case of shrimp Tpm, the coiled coil is a relative rigid and straight rod at both acidic and neutral pH across the 70 ns trajectory. In contrast, pig Tpm is significantly more dynamic, sampling a range of bent conformations. Significant unfolding is not seen on the timescales of these simulations, but larger global flexibility of pig Tpm may allow for transient unfolding, leading to proteolytic digestion. Overall, the molecular dynamics simulations point to a mechanism of epitope selection based on elevated flexibility of Ala-clusters, and allergen vs. non-allergen discrimination based on global structural flexibility.
Figure 7.
Molecular dynamics trajectories of shrimp (blue) and pig (gold) Tpm highlight enhanced global flexibility of vertebrate form. Radius of gyration, Rg, was calculated over Cα positions for each trajectory. See also Figures S6, S7.
We sought to understand allergenicity and epitope identity from a structural perspective using a simple coiled-coil allergen as a model protein. In vitro experiments showed that epitopes preferentially survive digestion. However, survival of the five epitopes utilized two strategies; high local stability of the two N terminal epitopes versus long range regional stability in the remaining C terminal epitopes. Epitope predictions using local dynamics calculated from MD simulations correctly identify epitopes in Tpm. Experimental and computational characterizations did not identify a single structural factor that would discriminate an allergen from a non-allergen, but significant differences in global dynamics may provide future insight. The correlations between digestibility, stability and food allergy in the case of Tpm are consistent with a more general tradeoff between stability and immunogenicity (Camacho et al., 2008). Understanding how structure and stability differentiate allergens will promote a better understanding of the immune interaction with antigens and inform protein engineering efforts to design protein therapeutics maximizing stability while minimizing immunogenicity (Griswold and Bailey-Kellogg, 2016; Schubert et al., 2018).
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by the Lead Contact Vikas Nanda (nanda@cabm.rutgers.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Tropomyosin proteins presented in this study were isolated from natural animal sources. Penaeus aztecus (brown shrimp) was purchased flash-frozen from a local supermarket and purified in the laboratory. Sus scrofa (pig) muscle tropomyosin was purchased from Sigma Aldrich (St. Louis) and subject to additional purification. Protein identity was verified by mass spectrometric analysis of coverage from LC-MS/MS.
METHOD DETAILS
Purification of shrimp tropomyosin (Pen a 1)
Approximately 400 g of brown shrimp (shelled and deveined) was minced then washed four times in 800 mL of a dilute salt solution [20 mM potassium chloride (KCl), 1.0 mM potassium bicarbonate, 0.1 mM calcium chloride, 0.1 mM dithiothreitol (DTT), pH 8.4; 5 min stir and filtered with cheese cloth each step]. The solid residue was washed three times in 1.0 L of 95% ethanol (10 min stir each step), then washed three times in 800 mL of chilled diethyl ether (5 min manual stir with glass rod). The solid residue was dried overnight at room temperature yielding 10 grams of dry protein powder, mixed overnight in 100 mL of extraction buffer [25 mM Tris, 1.0 M sodium chloride (NaCl), 2 mM EDTA, 1.0 mM DTT, pH 7.5], and centrifuged at 18,000 rpm for 40 min to recover the protein supernatant. To every 100 mL of protein solution recovered, 50 mL of 1 M NaCl was added and the mixture was stirred for two hours followed by the dropwise addition of glacial acetic acid to lower the pH to 4.5 for isoelectric precipitation. The protein suspension was centrifuged at 18,000 rpm for 40 min, and the pellet was dissolved in dialysis buffer (10 mM Tris, 100 mM NaCl, pH 7.5). The mixture was clarified by centrifugation (18,000 rpm for 40 min) followed by sequential protein precipitations at 25% and 40% ammonium sulfate. The protein pellet obtained after the 40% ammonium sulfate precipitation was dissolved in approximately 10 ml of dialysis buffer, and dialyzed with three buffer exchanges (1.5–2.0 L each exchange) over a period of 1 week. All washing, extraction and purification steps were performed at 4 °C.
Final purification was performed on an AKTA FPLC system (GE Healthcare Lifesciences) using a Superdex 200 10/300 GL column pre-equilibrated with wash buffer (10 mM Tris, 100 mM NaCl, pH 7.5) at 4 °C. A volume of 200 μl of Pen a 1 (5 mg/ml) was loaded on the column, and 0.5 ml fractions were collected at a flow rate of 0.7 mL/min. The absorbance spectrum of the elutant was monitored at 280 nm. Fractions containing purified protein were pooled and stored at −20 °C for later use.
Purification of porcine tropomyosin
Tropomyosin from porcine muscle was purchased (Sigma Chemicals, St Louis, MO) and reconstituted in ultrapure water. A volume of 200 μl (5 mg/ml) was purified on an AKTA FPLC system as previously described. Fractions containing purified protein were pooled and stored at −20 °C.
Characterization of peptide/protein by circular dichroism (CD)
The sample was mixed in either basic buffer (20 mM sodium phosphate, 150 mM NaCl, pH 8) or acidic buffer [20 mM sodium phosphate, 150 mM NaCl, pH 2 adjusted with 12 N hydrochloric acid (HCl)]. CD wavelength spectra were measured (190–260 nm scans, 6 sec averaging, 4 °C) on an AVIV model 420SF spect rophotometer (Aviv Biomedical, Lakewood, NJ). Blank buffer subtraction was performed on each sample and the molar residual ellipticity (MRE) was calculated from the sample concentration, the number of residues, and a cell path length of 0.1 cm. Thermal denaturation spectra of the samples were performed at 222 nm from 4–95°C (rate of 0.5°C /step, 0.5 min temperature equilibration time, 6 sec averaging time).
Simulated gastric digestion of tropomyosin
Full length tropomyosin (shrimp or pig) was incubated for one hour at 37 °C in acidic buffer [0.2 M HCl−potassium chloride (KCl), pH 2], followed by the addition of pepsin A (Worthington Biochemical Corp., Lakewood, NJ) to give 0.33 Units of pepsin activity/μg tropomyosin substrate. Aliquots of the mixture were quenched at various times (0, 5, 10, 15, 30, 60, 120 and 240 min) with 0.2 M sodium bicarbonate (final pH 8–9). Samples were stored at −20 °C for later analysis.
Simulated gastrointestinal digestion of tropomyosin
The pH of digested tropomyosin (60 and 120 min gastric digestion) from above was adjusted to a range of 7.5 −7.9 with 12 N HCl at 37 °C. Trypsin (0.20 Units activity/ μg total substrate) and chymotrypsin (0.04 Units activity/μg total substrate) were added to the mixture. Aliquots of the mixture were quenched at various times (0, 15 and 30 min) with 5 mM Pefabloc SC (Sigma Chemicals, St Louis, MO). Samples were stored at −20 °C for later analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)
Samples were prepared by mixing digested peptides/protein with an equal volume of Laemelli sample buffer (Bio-Rad, Hercules, CA) under reducing conditions with β-mercaptoethanol. The samples were heated at 95 °C f or 5 min, then run on a 16.5% precast Tris-tricine gel (Bio-Rad) and stained with silver nitrate (SilverQuestTM staining kit, Life Technologies, Carlsbad, CA). Polypeptide SDS-PAGE standard (Bio-Rad) was used for the protein MW standards ladder.
MALDI-TOF mass spectrometry
MALDI-TOF experiments and analysis were conducted at the Rutgers Biological Mass Spectrometry Facility. Samples for both shrimp and pig tropomyosin were prepared as described above, and subject to simulated gastric digestion in pepsin for 0, 10, 30, 60, 120 and 240 minutes. Subsequent intestinal digestion was performed for 30 minutes starting with gastric 60 or 120 digestion end-products. Samples were brought to a pH of 8–9 and final concentration of 0.33 mg/mL for 0, 10, 30 and 240 min gastric digestions, or 0.30 mg/mL for gastric/intestinal digestions of 60 or 120 minutes. Prior to analysis, samples were 1:5 diluted with matrix (10mg/ml sinapinic acid in 50% acetonitrile, 0.1% trifloroacetic acid) and deposited on an opti-TOF 384 well insert for MALDI-TOF/TOF (ABSciex) using dry-droplet method. The MALDI-TOF data were acquired using 4800 MALDI-TOF/TOF (ABSciex) with linear mid mass positive mode. Data were acquired from 3kDa to 40 kDa mass range with external calibration by apo-myoglobin singly charged, doubly charged ions as well as dimer ions. The laser was fixed at 5.6 kV, and spectra were based on accumulation of 1000 laser shots. Peaks with a minimum signal to noise ratio of 10 were exported from the Applied Biosystems 4000 Series database into mgf files using the TS2 Mascot utility.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS)
Peptides were solubilized in 0.1% trifluoroacetic acid, and analyzed by LC-MS/MS (Dionex Ultimate 3000 RLSCnano System interfaced with Velos LTQ Orbitrap (Thermo Scientific). Samples were separated by reverse phase HPCL (Q-C18, 2.1×50 mm, 3 um, 150 Å, CMP Scientific). After loading to the column, sample was washed with 2% loading Buffer B (A: A: 0.2% formic acid, B: 0.2% formic acid in acetonitrile) for 4 min with a flow rate of 200 μl/min and separated with a linear gradient of 2–50% B in 25 min before the column was washed with 90% B for 5 min and then equilibrated for 15 min with 2% B. Mass spectrometry data was acquired using a data-dependent acquisition procedure with a cyclic series of a full scan acquired from mass 300–2000 with resolution of 60,000 followed by MS/MS in the ion trap of the 20 most intense ions and a dynamic exclusion duration of 20 sec.
Peak lists in the format of MASCOT Generic Format (MGF.) was generated using the Proteome Discover 1.3 (ThermoFisher). Data were searched against custom database composed of more than 200 custom proteins including tropomyosin from pig and shrimp using a local version of the Global Proteome Machine (GPM) XE Manager version 2.2.1 (Beavis Informatics Ltd., Winnipeg, Canada) with X!Tandem version 10–12-01–1 to assign spectral data 1, 2. Precursor ion mass error tolerance was set to ±10 ppm and fragment mass error tolerance to ±0.4 Da. Protease specificity were set as non-specific. Methionine oxidation and deamidation at asparagine and glutamine residues were set as variable modifications with refinement of dioxidation on methionine, oxidation and dioxidation on tryptophan. All LC-MS data were analyzed together in a MudPit analysis. The resulting identifications were filtered by log GPM expectancy score (logE, <−5 for protein and <−2 for peptide).
Molecular Dynamics Simulations
The high resolution structure of the pig Tpm structure was used (PDB ID: 1C1G) (Whitby and Phillips, 2000). The shrimp homolog was derived from it by making the required substitutions using the protCAD (protein Computer Aided Design) software (Pike and Nanda, 2015; Summa, 2002). The pig sequence is from Sus scrofa (NCBI Reference Sequence: NP_001090952.1) and the shrimp from Penaeus monodon, identical to P. aztecus (Genbank: AAX37288.1). Proteins were solvated with a TIP3P water box with a distance of 10 Å from the protein, using the solvatebox radius command in AMBER (Case et al., 2014). The final box dimensions were 77.1 × 62.5 × 452.6 Å (54 Na+, 174312 waters) for pig Tpm and 116.6 × 64.7 × 417.7 Å (46 Na+, 262218 waters) for shrimp Tpm.
Structural models were then subjected to molecular dynamics simulations using AMBER14 (Case et al., 2014), under the ff14sb force field. The ion force field was derived from parameters of Joung and Cheatham (Joung and Cheatham, 2008, 2009) for ions in TIP3P water. Two rounds of optimization were performed to energetically minimize the structure, starting with the steepest descent method and then the conjugate gradient method. Molecular Dynamics was performed starting with a 2 fs time step required for the SHAKE algorithm to constrain bond lengths. Periodic boundaries were set under constant volume. The system was thermalized from 0 to 300 K using Langevin dynamics with a collision frequency of 3 ps−1. Next an equilibration step was run under constant temperature with no pressure scaling for 100 ps. The final MD trajectory was for 90 ns in the NVT ensemble using the weak-coupling algorithm (Berendsen et al., 1984), maintaining temperature at 300 K, using the Berendsen barostat for pressure scaling (reference pressure set to 1 bar, relaxation time 1.0 ps).
Long-range electrostatics were modeled using the particle mesh Ewald method (Darden et al., 1993), with non-bonded interaction cutoff set to 8.0 Å. Coordinates were recorded every 20 ps. Simulations were allowed to equilibrate for 20 ns, during which time the shrimp RMSD reached a consistent level (Fig. S6). Trajectories were analyzed from 20 – 90 ns in the trajectory with pytraj, a CPPTRAJ-based python module (Roe and Cheatham III, 2013), and MATLAB.
Peptide design, synthesis and purification
Peptides were synthesized at the Tufts University Core Facility (http://tucf.org) using solid-phase FMOC chemistry, purified by reverse-phase HPLC, and verified by mass spectrometry. N- and C- termini were acetylated and amidated, respectively. Peptides were dialyzed in filtered deionized water, lyophilized and stored at −20°C. Peptides were dissolved in 20 mM phosphate buffer, 150 mM NaCl, pH 8. Peptide concentrations were confirmed by measuring the absorbance at 214 nm, using an extinction coefficient of 2200 M−1 cm−1 per peptide bond on an AVIV model 14DS UV–Vis spectrophotometer. Peptide identifier, sequence, and corresponding Tpm residues are listed in Table S1.
QUANTIFICATION AND STATISTICAL ANALYSIS
All LC-MS data were analyzed together in a MudPit analysis. The resulting identifications were filtered by log GPM expectancy score (logE, <−5 for protein and <−2 for peptide).
DATA AND SOFTWARE AVAILABILITY
LC-MS/MS raw data sets and peptide analysis for gastric and intestinal digestion of shrimp and pig tropomyosin are available in the Peptide Atlas: www.peptideatlas.org under dataset identifier PASS01188.
Shrimp gastric digestion 0 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS433.raw
Shrimp gastric digestion 10 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS434.raw
Shrimp gastric digestion 30 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS435.raw
Shrimp gastric digestion 60 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS437.raw
Shrimp gastric digestion 120 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS439.raw
Shrimp gastric digestion 240 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS436.raw
Shrimp gastric digestion 60 minutes and intestinal digestion 30 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS438.raw
Shrimp gastric digestion 120 minutes and intestinal digestion 30 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS440.raw
Pig gastric digestion 0 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS441.raw
Pig gastric digestion 10 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS442.raw
Pig gastric digestion 30 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS443.raw
Pig gastric digestion 60 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS445.raw
Pig gastric digestion 120 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS447.raw
Pig gastric digestion 240 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS444.raw
Pig gastric digestion 60 minutes and intestinal digestion 30 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS446.raw
Pig gastric digestion 120 minutes and intestinal digestion 30 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS448.raw
LC-MS/MS peptide identification and analysis (source data for Figure 3): http://www.peptideatlas.org/PASS/PASS01188/LCMSMS_Shrimp_digestion_131003120210_shrimp_NEW.xls
http://www.peptideatlas.org/PASS/PASS01188/LCMSMS_Pig_digestion_131003164448_pig_NEW.xls
Supplementary Material
HIGHLIGHTS.
Sequence-based immunogenicity metrics do not discriminate allergen forms of Tpm
Tpm allergens are resistant to simulated gastric and intestinal digestion
Epitopes are more stable under gastric and intestinal conditions
Dynamics discriminate epitopes and allergen Tpms from non-allergens
ACKNOWLEDGEMENTS
We thank Sarah Hitchcock-DeGregori for helping develop the shrimp Tpm purification protocol, providing rat Tpm protein, and many useful scientific discussions. Supporting experiments and analysis were performed by Rashesh Shah, Paras Patel, Aisha Jasani and Rebekah Amarini. This work was supported by the National Institutes of Health R21 AI-088627.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Astwood JD, Leach JN, and Fuchs RL (1996). Stability of food allergens to digestion in vitro. Nat Biotechnol 14, 1269–1273. [DOI] [PubMed] [Google Scholar]
- Ayuso R, Lehrer SB, and Reese G (2002). Identification of continuous, allergenic regions of the major shrimp allergen Pen a 1 (tropomyosin). Int Arch Allergy Immunol 127, 27–37. [DOI] [PubMed] [Google Scholar]
- Ayuso R, Lehrer SB, Tanaka L, Ibanez MD, Pascual C, Burks AW, Sussman GL, Goldberg B, Lopez M, and Reese G (1999). IgE antibody response to vertebrate meat proteins including tropomyosin. Ann Allergy Asthma Immunol 83, 399–405. [DOI] [PubMed] [Google Scholar]
- Barua B, Pamula MC, and Hitchcock-DeGregori SE (2011). Evolutionarily conserved surface residues constitute actin binding sites of tropomyosin. Proc Natl Acad Sci U S A 108, 10150–10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen HJC, Postma JPM, Gunsteren W.F.v., DiNola A, and Haak JR (1984). Molecular dynamics with coupling to an external bath. The Journal of Chemical Physics 81, 3684–3690. [Google Scholar]
- Brown JH, Kim KH, Jun G, Greenfield NJ, Dominguez R, Volkmann N, Hitchcock-DeGregori SE, and Cohen C (2001). Deciphering the design of the tropomyosin molecule. Proc Natl Acad Sci U S A 98, 8496–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JH, Zhou Z, Reshetnikova L, Robinson H, Yammani RD, Tobacman LS, and Cohen C (2005). Structure of the mid-region of tropomyosin: bending and binding sites for actin. Proc Natl Acad Sci U S A 102, 18878–18883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho CJ, Katsumata Y, and Ascherman DP (2008). Structural and thermodynamic approach to peptide immunogenicity. PLoS Comput Biol 4, e1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilleri M, Colemont LJ, Phillips SF, Brown ML, Thomforde GM, Chapman N, and Zinsmeister AR (1989). Human gastric emptying and colonic filling of solids characterized by a new method. Am J Physiol 257, G284–290. [DOI] [PubMed] [Google Scholar]
- Case DA, Babin V, Berryman J, Betz R, Cai Q, Cerutti D, Cheatham Iii T, Darden T, Duke R, and Gohlke H (2014). Amber 14. [Google Scholar]
- Commission, J.F.W.C.A., Programme, J.F.W.F.S., and Organization, W.H. (2003). Codex Alimentarius: Food hygiene, basic texts (Food & Agriculture Org.). [Google Scholar]
- Darden T, York D, and Pedersen L (1993). Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. The Journal of Chemical Physics 98, 10089–10092. [Google Scholar]
- DeWitt AM, Mattsson L, Lauer I, Reese G, and Lidholm J (2004). Recombinant tropomyosin from Penaeus aztecus (rPen a 1) for measurement of specific immunoglobulin E antibodies relevant in food allergy to crustaceans and other invertebrates. Mol Nutr Food Res 48, 370–379. [DOI] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. (2014). Pfam: the protein families database. Nucleic Acids Research 42, D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzsimmons CM, and Dunne DW (2009). Survival of the fittest: allergology or parasitology? Trends Parasitol 25, 447–451. [DOI] [PubMed] [Google Scholar]
- Fu TJ, Abbott UR, and Hatzos C (2002). Digestibility of food allergens and nonallergenic proteins in simulated gastric fluid and simulated intestinal fluid-a comparative study. J Agric Food Chem 50, 7154–7160. [DOI] [PubMed] [Google Scholar]
- Geeves MA, Hitchcock-DeGregori SE, and Gunning PW (2015). A systematic nomenclature for mammalian tropomyosin isoforms. J Muscle Res Cell Motil 36, 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Fernandez J, Veleiro B, Daschner A, and Cuellar C (2016). Are fish tropomyosins allergens? Ann Allergy Asthma Immunol 116, 74–76 e75. [DOI] [PubMed] [Google Scholar]
- Greenfield NJ, Huang YJ, Palm T, Swapna GV, Monleon D, Montelione GT, and Hitchcock-DeGregori SE (2001). Solution NMR structure and folding dynamics of the N terminus of a rat non-muscle alpha-tropomyosin in an engineered chimeric protein. J Mol Biol 312, 833–847. [DOI] [PubMed] [Google Scholar]
- Griswold KE, and Bailey-Kellogg C (2016). Design and engineering of deimmunized biotherapeutics. Current Opinion in Structural Biology 39, 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbury PB, Zhang T, Kim PS, and Alber T (1993). A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262, 1401–1407. [DOI] [PubMed] [Google Scholar]
- Herman RA, Woolhiser MM, Ladics GS, Korjagin VA, Schafer BW, Storer NP, Green SB, and Kan L (2007). Stability of a set of allergens and non-allergens in simulated gastric fluid. Int J Food Sci Nutr 58, 125–141. [DOI] [PubMed] [Google Scholar]
- Hitchcock-DeGregori SE (2008). Tropomyosin: function follows structure. Adv Exp Med Biol 644, 60–72. [DOI] [PubMed] [Google Scholar]
- Ishii Y, Hitchcock-DeGregori S, Mabuchi K, and Lehrer SS (1992). Unfolding domains of recombinant fusion alpha alpha-tropomyosin. Protein Sci 1, 1319–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson BA, and Wolf H (1988). The antigenic index: a novel algorithm for predicting antigenic determinants. Comput Appl Biosci 4, 181–186. [DOI] [PubMed] [Google Scholar]
- Jenkins JA, Breiteneder H, and Mills EN (2007). Evolutionary distance from human homologs reflects allergenicity of animal food proteins. J Allergy Clin Immunol 120, 1399–1405. [DOI] [PubMed] [Google Scholar]
- Joung IS, and Cheatham TE (2008). Determination of Alkali and Halide Monovalent Ion Parameters for Use in Explicitly Solvated Biomolecular Simulations. The Journal of Physical Chemistry B 112, 9020–9041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung IS, and Cheatham TE (2009). Molecular Dynamics Simulations of the Dynamic and Energetic Properties of Alkali and Halide Ions Using Water-Model-Specific Ion Parameters. The Journal of Physical Chemistry B 113, 13279–13290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan IJ, Di R, Patel P, and Nanda V (2013). Evaluating pH-induced gastrointestinal aggregation of Arachis hypogaea 1 fragments as potential components of peanut allergy. J Agric Food Chem 61, 8430–8435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladics GS, Cressman RF, Herouet-Guicheney C, Herman RA, Privalle L, Song P, Ward JM, and McClain S (2011). Bioinformatics and the allergy assessment of agricultural biotechnology products: industry practices and recommendations. Regul Toxicol Pharmacol 60, 46–53. [DOI] [PubMed] [Google Scholar]
- Lehrer SS, and Yuan A (1998). The Stability of Tropomyosin at Acid pH: Effects of Anion Binding. Journal of Structural Biology 122, 176–179. [DOI] [PubMed] [Google Scholar]
- Li Y, Mui S, Brown JH, Strand J, Reshetnikova L, Tobacman LS, and Cohen C (2002). The crystal structure of the C-terminal fragment of striated-muscle alphatropomyosin reveals a key troponin T recognition site. Proc Natl Acad Sci U S A 99, 7378–7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren DH, Hwang SI, Wu L, and Han DK (2010). Role of spectral counting in quantitative proteomics. Expert Rev Proteomics 7, 39–53. [DOI] [PubMed] [Google Scholar]
- Lupas AN, and Gruber M (2005). The structure of alpha-helical coiled coils. Adv Protein Chem 70, 37–78. [DOI] [PubMed] [Google Scholar]
- Ly S, and Lehrer SS (2012). Long-range effects of familial hypertrophic cardiomyopathy mutations E180G and D175N on the properties of tropomyosin. Biochemistry 51, 6413–6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleki SJ, Viquez O, Jacks T, Dodo H, Champagne ET, Chung SY, and Landry SJ (2003). The major peanut allergen, Ara h 2, functions as a trypsin inhibitor, and roasting enhances this function. J Allergy Clin Immunol 112, 190–195. [DOI] [PubMed] [Google Scholar]
- Matyushenko AM, Artemova NV, Sluchanko NN, and Levitsky DI (2015). Effects of two stabilizing substitutions, D137L and G126R, in the middle part of alphatropomyosin on the domain structure of its molecule. Biophys Chem 196, 77–85. [DOI] [PubMed] [Google Scholar]
- McLachlan AD, Stewart M, and Smillie LB (1975). Sequence repeats in alphatropomyosin. J Mol Biol 98, 281–291. [DOI] [PubMed] [Google Scholar]
- Murakami K, Stewart M, Nozawa K, Tomii K, Kudou N, Igarashi N, Shirakihara Y, Wakatsuki S, Yasunaga T, and Wakabayashi T (2008). Structural basis for tropomyosin overlap in thin (actin) filaments and the generation of a molecular swivel by troponin-T. Proc Natl Acad Sci U S A 105, 7200–7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevzorov IA, Nikolaeva OP, Kainov YA, Redwood CS, and Levitsky DI (2011). Conserved noncanonical residue Gly-126 confers instability to the middle part of the tropomyosin molecule. J Biol Chem 286, 15766–15772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea EK, Klemm JD, Kim PS, and Alber T (1991). X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science 254, 539–544. [DOI] [PubMed] [Google Scholar]
- Ohgushi M, and Wada A (1983). ‘Molten-globule state’: a compact form of globular proteins with mobile side-chains. FEBS Lett 164, 21–24. [DOI] [PubMed] [Google Scholar]
- Pali-Scholl I, and Jensen-Jarolim E (2011). Anti-acid medication as a risk factor for food allergy. Allergy 66, 469–477. [DOI] [PubMed] [Google Scholar]
- Park C, and Marqusee S (2004). Probing the high energy states in proteins by proteolysis. J Mol Biol 343, 1467–1476. [DOI] [PubMed] [Google Scholar]
- Pike DH, and Nanda V (2015). Empirical estimation of local dielectric constants: Toward atomistic design of collagen mimetic peptides. Peptide Science 104, 360–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potekhin SA, and Privalov PL (1982). Co-operative blocks in tropomyosin. J Mol Biol 159, 519–535. [DOI] [PubMed] [Google Scholar]
- Reese G, Ayuso R, and Lehrer SB (1999). Tropomyosin: an invertebrate pan-allergen. Int Arch Allergy Immunol 119, 247–258. [DOI] [PubMed] [Google Scholar]
- Reese G, Ayuso R, Leong-Kee SM, Plante M, and Lehrer SB (2001). The IgE-binding regions of the major allergen Pen a 1: Multiple epitopes or intramolecular cross-reactivity? Int Arch Allergy Imm 124, 103–106. [Google Scholar]
- Reese G, Viebranz J, Leong-Kee SM, Plante M, Lauer I, Randow S, Moncin MS, Ayuso R, Lehrer SB, and Vieths S (2005). Reduced allergenic potency of VR9–1, a mutant of the major shrimp allergen Pen a 1 (tropomyosin). J Immunol 175, 8354–8364. [DOI] [PubMed] [Google Scholar]
- Roe DR, and Cheatham TE III (2013). PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. Journal of chemical theory and computation 9, 3084–3095. [DOI] [PubMed] [Google Scholar]
- Sampson HA (1999). Food allergy. Part 1: immunopathogenesis and clinical disorders. J Allergy Clin Immunol 103, 717–728. [DOI] [PubMed] [Google Scholar]
- Schubert B, Scharfe C, Donnes P, Hopf T, Marks D, and Kohlbacher O (2018). Population-specific design of de-immunized protein biotherapeutics. PLoS Comput Biol 14, e1005983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicherer SH, Munoz-Furlong A, and Sampson HA (2004). Prevalence of seafood allergy in the United States determined by a random telephone survey. J Allergy Clin Immunol 114, 159–165. [DOI] [PubMed] [Google Scholar]
- Sicherer SH, and Sampson HA (2010). Food allergy. J Allergy Clin Immunol 125, S116–125. [DOI] [PubMed] [Google Scholar]
- Singh A, and Hitchcock-DeGregori SE (2003). Local destabilization of the tropomyosin coiled coil gives the molecular flexibility required for actin binding. Biochemistry 42, 14114–14121. [DOI] [PubMed] [Google Scholar]
- Singh A, and Hitchcock-DeGregori SE (2006). Dual requirement for flexibility and specificity for binding of the coiled-coil tropomyosin to its target, actin. Structure 14, 43–50. [DOI] [PubMed] [Google Scholar]
- Sumida JP, Wu E, and Lehrer SS (2008). Conserved Asp-137 imparts flexibility to tropomyosin and affects function. J Biol Chem 283, 6728–6734. [DOI] [PubMed] [Google Scholar]
- Summa CM (2002). protCAD (University of Pennsylvania School of Medicine: Philadelphia, PA: ). [Google Scholar]
- Talley K, and Alexov E (2010). On the pH-optimum of activity and stability of proteins. Proteins 78, 2699–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SL (1997). Food from genetically modified organisms and potential for food allergy. Environ Toxicol Pharmacol 4, 121–126. [DOI] [PubMed] [Google Scholar]
- Taylor SL (2002). Protein allergenicity assessment of foods produced through agricultural biotechnology. Annu Rev Pharmacol 42, 99–112. [DOI] [PubMed] [Google Scholar]
- Tyagi N, Farnell EJ, Fitzsimmons CM, Ryan S, Tukahebwa E, Maizels RM, Dunne DW, Thornton JM, and Furnham N (2015). Comparisons of Allergenic and Metazoan Parasite Proteins: Allergy the Price of Immunity. PLoS Comput Biol 11, e1004546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untersmayr E, Scholl I, Swoboda I, Beil WJ, Forster-Waldl E, Walter F, Riemer A, Kraml G, Kinaciyan T, Spitzauer S, et al. (2003). Antacid medication inhibits digestion of dietary proteins and causes food allergy: a fish allergy model in BALB/c mice. J Allergy Clin Immunol 112, 616–623. [DOI] [PubMed] [Google Scholar]
- Whitby FG, and Phillips GN Jr. (2000). Crystal structure of tropomyosin at 7 Angstroms resolution. Proteins 38, 49–59. [PubMed] [Google Scholar]
- Williams DL Jr., and Swenson CA (1981). Tropomyosin stability: assignment of thermally induced conformational transitions to separate regions of the molecule. Biochemistry 20, 3856–3864. [DOI] [PubMed] [Google Scholar]
- Woods EF (1977). Stability of segments of rabbit alpha-tropomyosin. Aust J Biol Sci 30, 527–542. [DOI] [PubMed] [Google Scholar]
- Zeng X, Herndon AM, and Hu JC (1997). Buried asparagines determine the dimerization specificities of leucine zipper mutants. Proc Natl Acad Sci U S A 94, 3673–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Hitchcock-DeGregori SE, and Barua B (2016). Investigating the effects of tropomyosin mutations on its flexibility and interactions with filamentous actin using molecular dynamics simulation. J Muscle Res Cell Motil 37, 131–147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
LC-MS/MS raw data sets and peptide analysis for gastric and intestinal digestion of shrimp and pig tropomyosin are available in the Peptide Atlas: www.peptideatlas.org under dataset identifier PASS01188.
Shrimp gastric digestion 0 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS433.raw
Shrimp gastric digestion 10 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS434.raw
Shrimp gastric digestion 30 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS435.raw
Shrimp gastric digestion 60 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS437.raw
Shrimp gastric digestion 120 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS439.raw
Shrimp gastric digestion 240 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS436.raw
Shrimp gastric digestion 60 minutes and intestinal digestion 30 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS438.raw
Shrimp gastric digestion 120 minutes and intestinal digestion 30 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS440.raw
Pig gastric digestion 0 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS441.raw
Pig gastric digestion 10 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS442.raw
Pig gastric digestion 30 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS443.raw
Pig gastric digestion 60 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS445.raw
Pig gastric digestion 120 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS447.raw
Pig gastric digestion 240 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS444.raw
Pig gastric digestion 60 minutes and intestinal digestion 30 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS446.raw
Pig gastric digestion 120 minutes and intestinal digestion 30 minutes: http://www.peptideatlas.org/PASS/PASS01188/VLS448.raw
LC-MS/MS peptide identification and analysis (source data for Figure 3): http://www.peptideatlas.org/PASS/PASS01188/LCMSMS_Shrimp_digestion_131003120210_shrimp_NEW.xls
http://www.peptideatlas.org/PASS/PASS01188/LCMSMS_Pig_digestion_131003164448_pig_NEW.xls