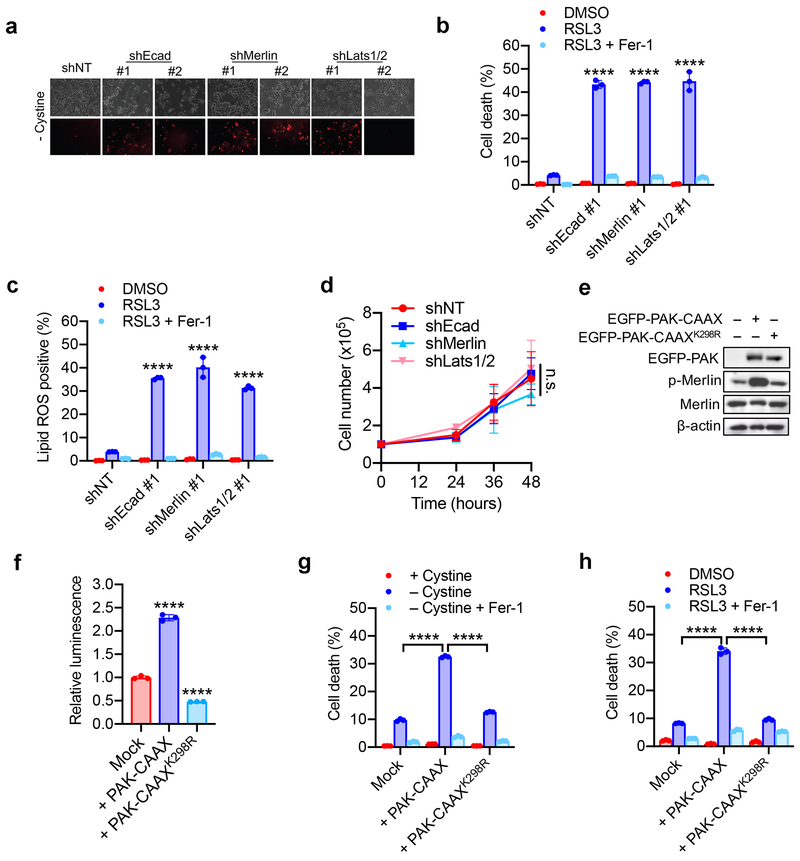
Extended Data Figure 4. The Hippo pathway links cell density and intercellular contact to ferroptosis.
a. Confluent cells were subjected to cystine starvation for 30 h. Dead cells were stained by PI.
b. HCT116 cells expressing shNT, shEcad, shMerlin, or shLats1/2 as indicated were treated with 5 µM RSL3 with or without 2 µM Fer-1. Cell death was measured at 18 h. Data are plotted as mean ± s.d.; n = 3 biological replicates. P-values were acquired by one-way ANOVA, **** p<0.0001.
c. Cells were treated as described in (b). Lipid ROS was assessed at 12 h after treatment. Data are plotted as mean ± s.d.; n = 3 biological replicates. P-values were acquired by one-way ANOVA, **** p<0.0001.
d. Cumulative cell growth curve expressed as the total cell count of shEcad, shMerlin and shLats1/2 HCT116 cells. Data are plotted as mean ± s.d.; n = 3 biological replicates. P-value was acquired by two-way ANOVA, n.s., p=0.9497.
e. HCT116 cells were transfected with EGFP-PAK containing a prenylation (CAAX) motif (thus constitutively active), or an inactive mutant form of PAK (K298R). Expression and phosphorylation of Merlin was assayed by Western blot.
f. HCT116 cells expressing activated or inactive PAK were transfected with the 8xGTIIC-luciferase reporter, and YAP/TEAD activity was measured by luminescence. Data are plotted as mean ± s.d.; n = 3 biological replicates. P-values were acquired by one-way ANOVA, **** p<0.0001.
g. HCT116 cells expressing activated or inactive PAK were plated at high density and treated with cystine-free media with or without 1 µM Ferrostatin-1 for 30 h. Data are plotted as mean ± s.d.; n = 3 biological replicates. P-values were acquired by one-way ANOVA, **** p<0.0001.
h. Cells were prepared as described in (g) and treated with DMSO or 5 µM RSL3 with or without 1 µM Ferrostatin-1. Cell death was measured at 24 h. Data are plotted as mean ± s.d.; n = 3 biological replicates. P-values were acquired by one-way ANOVA, **** p<0.0001.