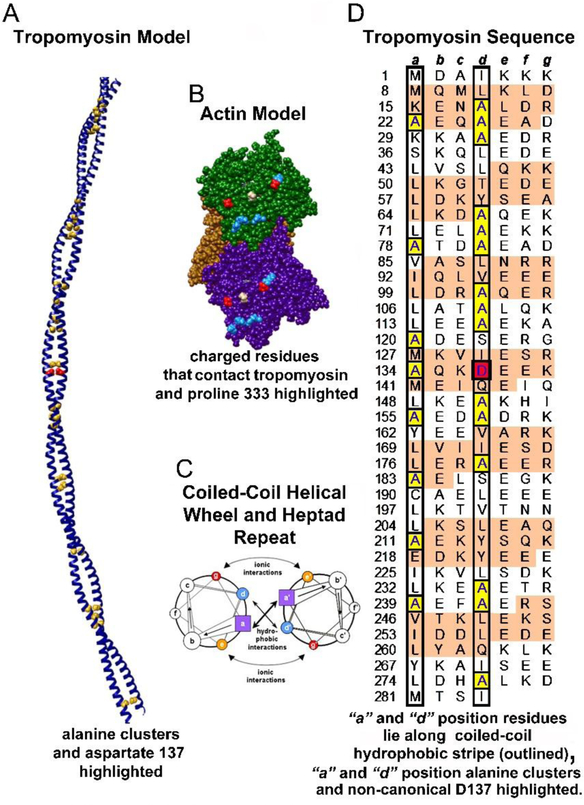
Figure 1. Tropomyosin architecture.
A. Ribbon diagram of the tropomyosin coiled coil with core alanines and aspartate-137 highlighted as yellow and red spheres respectively. B. Three neighboring actin subunits of F-actin highlighting charged residues (D25 and D311 in red and R28, R147, K315, K326 and K328 in blue). These residues are likely to interact electrostatically with successive pseudo-repeats of tropomyosin. Proline 333 (tan), which forms the boundary between blocked and closed states, is also highlighted. C. Helical wheel diagram of a canonical dimeric coiled-coil. Heptad positions are labeled a to g and a’ to g’ for the respective helices of the dimer; figure adapted from Hagemann et al (2008). D. Striated muscle tropomyosin (Tpm1.1) sequence annotation; figure adapted and modified from Brown and Cohen (2005) and Lehman et al (2018) to emphasize residues within heptad repeats (a, b, c, d, e, f, g) in each chain of the tropomyosin coiled-coil homodimer, with the first residue in each heptad repeat numbered. The α-zones (as defined in Brown and Cohen (2005)) of each pseudo-repeat of tropomyosin that locate close to actin subdomains 1 and 3 are shaded in tan, while the β-zones found over subdomains 2 and 4 have a white background (Brown and Cohen 2005). Generally, hydrophobic residues are localized at a and d residue positions (found within black borders in the diagram). Core alanine residues are highlighted in yellow and the d-position aspartate-137 is highlighted in red.