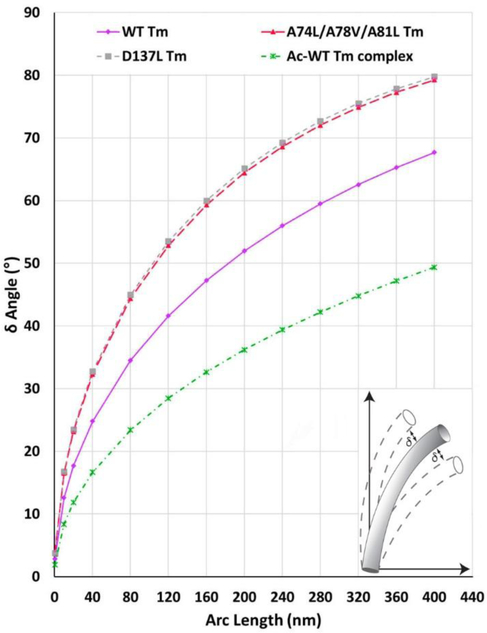
Figure 3. Tropomyosin flexibility.
The magnitude of isolated tropomyosin’s bending fluctuations during molecular dynamics simulations. Tropomyosin flexibility was assessed during MD by comparing the angular deflection (δ) of different length segments of individual tropomyosin coiled-coil conformers with their average conformation (Li et al 2010; Moore et al 2011) (see inset schematically illustrating δ for single conformers). The extent of the angular deflections along tropomyosin will increase exponentially at increasing distance from a fixed point. In the plots presented, normalized bending fluctuations of tropomyosin at any arc length away from a fixed displacement (0 nm on plot) are defined by the relationship, , where L is segment length and P is the persistence length of either control or mutant tropomyosins (Table 1). Displayed are plots of wildtype tropomyosin (magenta circles) as well as mutant D137L and A74L/A78V/A81L tropomyosins (grey square and red triangles, respectively). Note the increased slope of the mutant tracings indicating enhanced average flexibility compared to wildtype. For example, at a distance of 20 nm from the origin reference point, mutant tropomyosins fluctuate by almost as much as wildtype tropomyosin at 40 nm. Also note that once linked to F-actin, tropomyosin fluctuations in the blocked-state configuration (green) are, as expected, diminished. In this case, δ, was calculated from MD simulations of the actin-tropomyosin complex (Li et al 2011; Lehman et al 2018).