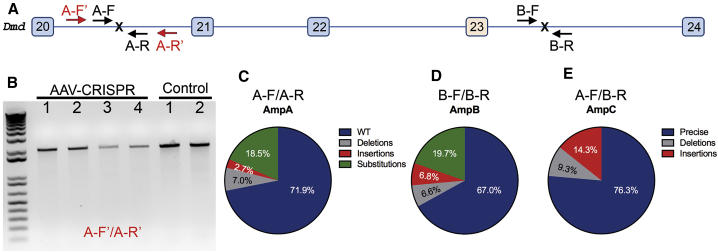
Figure 3.
CRISPR-Mediated DSBs at Dystrophin Locus in Dystrophic Mouse Heart Were Repaired Mainly by Precise Ligation of the Two Cut Ends
(A) Scheme illustrating the primers for PCR to detect larger deletions at the intron 20 target site (A-F′ and A-R′, 2 kbp), and for targeted deep-sequencing analysis (amplicon A, A-F/A-R; amplicon B, B-F/B-R; and amplicon C, A-F/B-R, 150 bp each). Since the unedited locus from A-F to B-R is over 23 kbp, PCR with A-F/B-R would not amplify the unedited DNA, while the correctly edited DNA would result in amplification of an ∼150-bp product. (B) Genomic DNA PCR showed only the expected 2 kbp product without smaller bands, indicating that no larger deletions at the target site were detected. (C–E) Targeted deep sequencing analyses of the amplicon A (C), B (D), and C (E) from pooled heart samples of five mdx mice treated with rAAV-CRISPR.