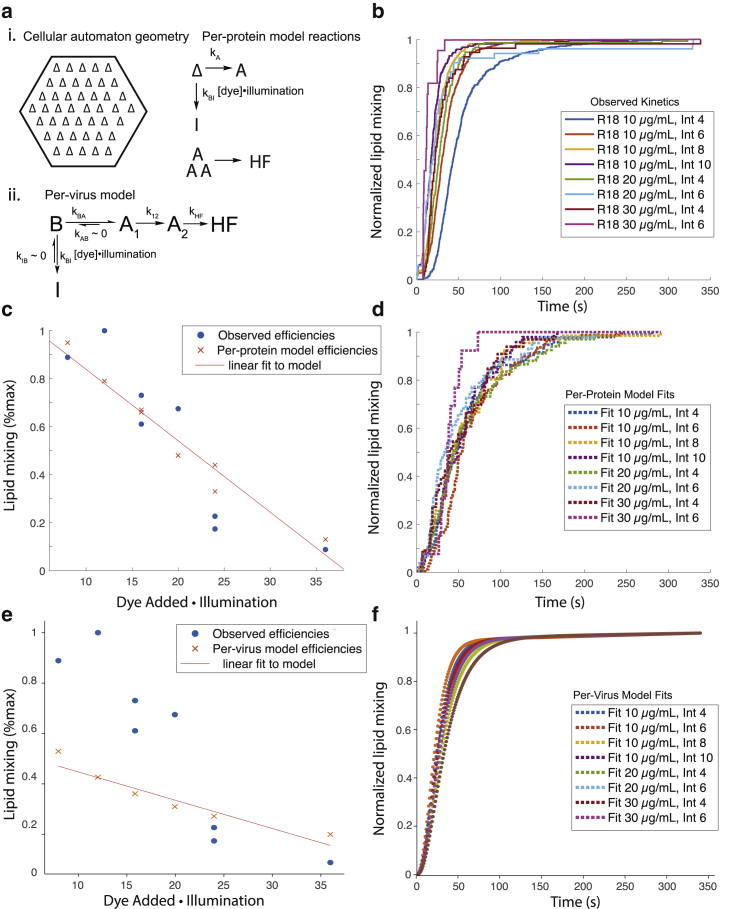
Figure 5.
Kinetic models of fusion protein inactivation can account for photoinactivation of fusion. Computational models of per-fusion-protein inactivation and per-virus inactivation (schematized in a) were fitted to single-virus fusion data at multiple R18 dye concentrations and illumination intensities (cumulative distribution functions in b). The models specify interconversion rates among bound (B), activated (A), inactivated (I), and hemifused (HF) states. In the per-protein model, states describe individual hemagglutinin trimers (except HF), whereas in the per-virus model, states describe the virus as a whole. The per-protein model could better account for the reduced fusion efficiency than the per-virus model (c and e) when lipid mixing was normalized across conditions. Cumulative distribution functions of modeled and observed fusion events normalized within each dye and illumination condition (b, d, and f) show that neither model robustly reproduces the slight speeding of fusion at increased dye illumination. Total numbers of lipid-mixing events used to construct the cumulative distribution events were 482, 677, 294, 222, 153, 52, 56, and 22 for the eight conditions plotted. The cellular automaton used in the model represents the virus:target membrane contact zone as a hexagonal lattice with individual hemagglutinin trimers that can convert to A or I. Three neighboring activated trimers result in HF. Legends in (b), (d), and (f) specify dye loading concentration and intensity (out of 255). To see this figure in color, go online.